From the desktop to the grid: Scalable bioinformatics via workflow conversion
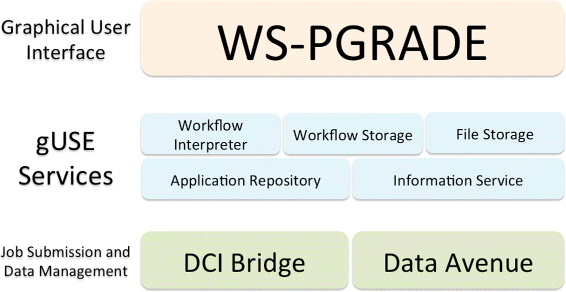 This featured article from the journal BMC Bioinformaticsfalls on the heels of several years of discussion on the topic of reproducibility of a scientific experiment's end results. De la Garza et al. point to workflows and their repeatability as vital cogs in such efforts. "Breaking down the complexity of such experiments into the joint collaboration of small, repeatable, well defined tasks, each with well defined inputs, parameters, and outputs, offers the immediate benefit of identifying bottlenecks, pinpoint sections which could benefit from parallelization," they state. The researchers developed their own set of free platform-independent tools for designing, executing, and sharing workflows. They conclude: "We are confident that our work presented in this document … not only provides scientists a way to design and test workflows on their desktop computers, but also enables them to use powerful resources to execute their workflows, thus producing scientific results in a timely manner."
This featured article from the journal BMC Bioinformaticsfalls on the heels of several years of discussion on the topic of reproducibility of a scientific experiment's end results. De la Garza et al. point to workflows and their repeatability as vital cogs in such efforts. "Breaking down the complexity of such experiments into the joint collaboration of small, repeatable, well defined tasks, each with well defined inputs, parameters, and outputs, offers the immediate benefit of identifying bottlenecks, pinpoint sections which could benefit from parallelization," they state. The researchers developed their own set of free platform-independent tools for designing, executing, and sharing workflows. They conclude: "We are confident that our work presented in this document … not only provides scientists a way to design and test workflows on their desktop computers, but also enables them to use powerful resources to execute their workflows, thus producing scientific results in a timely manner."
















