Information management for enabling systems medicine
| Full article title | Information management for enabling systems medicine |
|---|---|
| Journal | Current Directions in Biomedical Engineering |
| Author(s) | Ganzinger, Matthias; Knaup, Petra |
| Author affiliation(s) | Heidelberg University's Institute of Medical Biometry and Informatics |
| Primary contact | Email: matthias dot ganzinger at med dot uni-heidelberg dot de |
| Year published | 2017 |
| Volume and issue | 3 (2) |
| Page(s) | 0105 |
| DOI | 10.1515/cdbme-2017-0105 |
| ISSN | 2364-5504 |
| Distribution license | Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International |
| Website | https://www.degruyter.com/view/j/cdbme.2017.3.issue-2/cdbme-2017-0105/cdbme-2017-0105.xml |
| Download | https://www.degruyter.com/downloadpdf/j/cdbme.2017.3.issue-2/cdbme-2017-0105/cdbme-2017-0105.xml (PDF) |
Abstract
Systems medicine is a data-oriented approach in research and clinical practice to support the study and treatment of complex diseases. It relies on well-defined information management processes providing comprehensive and up-to-date information as the basis for electronic decision support. The authors suggest a three-layer information technology (IT) architecture for systems medicine and a cyclic data management approach, including a knowledge base that is dynamically updated by extract, transform, and load (ETL) procedures. Decision support is suggested as case-based and rule-based components. Results are presented via a user interface to acknowledging clinical requirements in terms of time and complexity. The systems medicine application was implemented as a prototype.
Keywords: systems medicine, information management, decision support systems
Introduction
Systems medicine is a current approach to aid physicians and researchers in the treatment and investigation of complex diseases. According to the definition of the European Commission, “‘Systems medicine‘ is the application of systems biology approaches to medical research and medical practice. Its objective is to integrate a variety of biological/medical data at all relevant levels of cellular organization using the power of computational and mathematical modelling, to enable understanding of the pathophysiological mechanisms, prognosis, diagnosis and treatment of disease.“[1] Consequently, the management of data is of great importance for systems medicine in research as well as clinical practice. Typically, data of different sources such as electronic health record systems, clinical research databases, or biomedical knowledge representations like ontologies have to be reviewed and prepared. The most prevalent data sources in systems medicine research projects are omics data and clinical data.[2]
Due to the comprehensive approach of systems medicine, neither disease-specific knowledge nor clinical data can be considered static. Thus, we suggest understanding information management for systems medicine as a dynamic process that evolves over time and leads to cyclic updates of the knowledge and data repositories behind the corresponding information technology (IT) system.
Further challenges arise from the broad availability of so-called omics data. This class of data — for example RNA microarray data — is characterized by a huge amount of attributes per sample that is often disproportional to the number of available cases. Currently, specific data preparation pipelines using statistical approaches like feature selection are necessary to make these data accessible for decision support solutions.[3]
Methods
For successful information management in the context of systems medicine, it is useful to distinguish between the IT architecture and the data management process. The architecture depends on the requirements of a specific systems medicine application. As a generic high-level architecture we propose a three-layer model[4]:
1. Data representation: Data and knowledge from different sources have to be prepared and made available for use in systems medicine. This includes data harmonization, transformation, and storage.
2. Decision support: Data and knowledge from layer 1 are processed by applying decision support approaches like case-based reasoning (CBR), deductive classifiers (rules-based), or systems biology models. Depending on the context, such components can be combined into hybrid systems.
3. User interface: Systems medicine applications should be designed to assist and not replace human decisions. Consequently, the user interface for such an application must be carefully designed to support well-informed, reproducible clinical decisions in an appropriate time frame.
The complexity of the data management process depends on the level of heterogeneity prevalent in the data sources. To achieve sufficient case numbers, it is often necessary to combine data on the same entity types from different sources. For example, hospitals may decide to collaborate and share clinical data on a specific disease area to build a joint systems medicine application with a higher number of cases and therefore greater statistical power (multi-center approach).
In most cases, clinical documentation will not be based on identical specifications. Thus, in a harmonization step, data definitions have to be evaluated for each attribute, both on a syntactic and semantic level. The resulting common data definition should be implemented into an automated extract, transform, and load (ETL) process to facilitate repeated loading of data to keep an up-to-date decision support system.[5][6]
Results
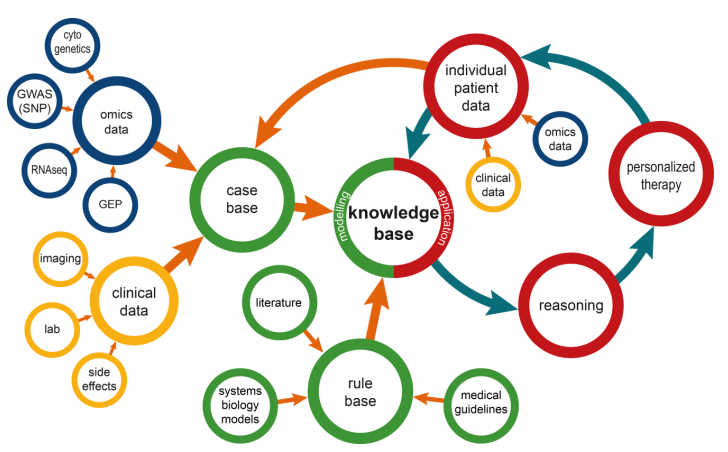
An overview of the resulting information management modelis shown in Figure 1. In the following paragraphs the elements of this model are described in detail.
|
Knowledge base
The core concept of the model is the knowledge base, which contains patient and disease related data, as well as formally represented knowledge. As such, it forms a systems medicine model in a broader sense. Specifically, the knowledge base is comprised of a case base and a rule base.
Case base
The case base covers information on the available experience in treating patients with a specific disease. Typically, such information is organized as case descriptions. Each case is described by a harmonized set of attributes covering clinical data, omics data, and others. The case base can be used for various research purposes like data mining or construction of systems biology models. In addition, its cases can be used for decision support directly by using the concept of patient similarity.[7] In the field of clinical artificial intelligence systems, patient similarity has been the subject of research for many years, most notably in case-based reasoning.[8]
In terms of clinical data, the case base contains typical data like diagnoses, procedures, side effects, and laboratory values. For some diseases, medical images such as computer tomography, magnetic resonance imaging, or microscope images can be included and be processed with corresponding similarity measures. More recently, case bases are enriched by molecular data describing different steps of the genetic process chain from DNA over proteins to cell-level regulatory processes.[3] However, the use of these data in the context of patient similarity is still challenging due to the large number of parameters involved.
Rule base
Since the case base only covers information on an institution’s previous experiences in treating a disease, it might not be comprehensive in terms of current evidence-based medical knowledge. This can be mitigated by adding a rule base to the systems medicine application. Rules formally represent medical knowledge in a way that can be interpreted by a rules engine like HertmiT.[9] Rules can be derived from various sources; medical treatment guidelines are rule sets intended for human interpretation that can be computerized. New findings on the treatment of a disease can be extracted from textual scientific literature by a manual or automated curation process.[10]
More suitable are sources that are computer-interpretable by design like gene ontology.[11] Published systems biology models can be part of a rule base in a broader sense since they provide machine-interpretable models, possibly described in systems biology mark-up language (SBML) and processed by a SBML simulation engine like COPASI.[12][13] For a rule base, a continuous curation process has to be established to ensure the timely availability of new knowledge, for example, when new treatment guidelines are published.
Inference
In contrast to systems biology, where understanding and in silico simulation of biological processes down to the cellular level are in focus, systems medicine always aims at supporting treatment decisions for individual patients. As shown in Figure 1, the knowledge base is the foundation for drawing conclusions for these individual patients. The technical aspects of this inference process differ in accordance with the type of knowledge available. For similarity-based inference methods like CBR, individual case instances are retrieved with a focus on maximizing similarity with the newly presented patient case. Consequently, a new patient has to be described using as many attributes as possible from the set of attributes in the case base. An individual treatment decision is made based on the outcome of the most similar patient from the case base. Especially for life threatening diseases like cancers, only the first treatment approach might be of interest for a newly diagnosed patient since later therapies cannot be considered independent of previous attempts.
For rule-based decision support, clinical data of new patients have to be defined in a way comparable to the case-based approaches. This set of individual attributes is used as input for the rules engine or model simulation. The result is a personalized treatment recommendation for the patient.
No matter which decision support method was used, the outcome of the treatment should be documented and added to the knowledge base, either as an additional case or by refining the rules and models as the patient population grows.
Prototype implementation
For the systems medicine project “clinically applicable, omics-based assessment of survival, side effects, and targets in multiple myeloma” (CLIOMMICS), a prototype of an IT system for systems medicine is being established according to the proposed architecture and data management process. As a disease model the project examines the multiple myeloma, a malignancy of plasma cells in the bone marrow.
Data (clinical parameters and omics data) have been harmonized and stored in a research data warehouse based on the open-source software “Informatics for Integrating Biology & the Bedside” (i2b2).[14] Data harmonization rules are documented as metadata which are used in an automated ETL process to ensure continuous updates of the case base [5]. Data in i2b2 are organized according to the star schema. While this data schema is optimized for analytical queries, for some purposes a flat case-oriented presentation of the data is desirable. We implemented a Generic Case Extractor (GCE) allowing a comprehensive data export as a matrix containing one line per case.[15]
While i2b2 can be used directly through its user interface for research purposes, it is also used as unified source for a case base. On this foundation, a case-based reasoning module was established with help of the Java-based CBR software framework myCBR.[16] To reflect the specific requirements of the cancer, a specific similarity measure based on survival data was developed. The user interface in form of a web portal with dedicated portlets visualizing CBR results is currently being developed. An additional part of the user interface is a report generator for generating medical letters covering results, e.g., for gene expression data.
Discussion
Information management for systems medicine is a demanding task requiring a multi-level approach to build a sustainable infrastructure. Special care has to be taken to address inherent dynamics of data that are used for systems medicine; over time the number of available health records will increase and treatment approaches will change, for example with the availability of new compounds. Such effects will have to be reflected in the corresponding knowledge base, no matter whether a case-based, rule-based, or other concept is implemented. In the authors’ opinion, the effort of harmonizing data for use in systems medicine should not only be used as a basis for clinical decision support but also made available for research (e.g., data mining). One possibility is the establishment of ETL processes for a biomedical data warehouse as suggested in this manuscript.
Evaluating a systems medicine application is challenging, especially in context of cancer diseases. Common in silico evaluation approaches like splitting patient data sets into training and test cohorts might not be applicable since it is hard to draw conclusions on test patients, who actually received a different treatment than the one the decision support component suggests. Eventually, a prospective controlled clinical trial might be necessary to compare the performance of a systems medicine application against unsupported decision making. However, such a trial will have to pass high ethical barriers.
Further research is necessary in the field of human-computer interaction. This is especially important for the field of systems medicine since physicians bear the burden of being responsible for the patient but only have limited resources in terms of time and budget at their disposal. Thus, it is necessary to provide the essence of a complex data analysis process to empower them to make a good decision for the health of patients.
The data management model presented here provides a blueprint for building comprehensive knowledge bases as they are required for systems medicine applications. Due to its generic nature, the model can be used with other IT systems as well.
Author's statement
Research funding: This work was funded by the German Ministry of Education and Research via the e:Med project CLIOMMICS (grant id: 01ZX1609A).
Conflict of interest: Authors state no conflict of interest.
Informed consent: Informed consent is not applicable.
Ethical approval: The conducted research is not related to either human or animals use.
References
- ↑ Auffray, C.; Balling, R.; Bensen, M. et al. (15 June 2010). "From Systems Biology to Systems Medicine". In Kyriakopoulou, C.; Mulligan, B. (PDF). European Commission. http://ec.europa.eu/research/health/pdf/systems-medicine-workshop-report_en.pdf.
- ↑ Gietzelt, M.; Löpprich, M.; Karmen, C. et al. (2016). "Models and Data Sources Used in Systems Medicine: A Systematic Literature Review". Methods of Information in Medicine 55 (2): 107–13. doi:10.3414/ME15-01-0151. PMID 26846174.
- ↑ 3.0 3.1 Anaissi, A.; Goyal, M.; Catchpoole, D.R. et al. (2015). "Case-based retrieval framework for gene expression data". Cancer Informatics 14: 21–31. doi:10.4137/CIN.S22371. PMC PMC4368049. PMID 25861214. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4368049.
- ↑ Ganzinger, M.; Gietzelt, M.; Karmen, C. et al. (2015). "An IT Architecture for Systems Medicine". Studies in Health Technology and Informatics 210: 185-9. PMID 25991127.
- ↑ Firnkorn, D.; Ganzinger, M.; Muley, T. et al. (2015). "A Generic Data Harmonization Process for Cross-linked Research and Network Interaction. Construction and Application for the Lung Cancer Phenotype Database of the German Center for Lung Research". Methods of Information in Medicine 54 (5): 455-60. doi:10.3414/ME14-02-0030. PMID 26394900.
- ↑ Vassiliadis, P. (2009). "A Survey of Extract–Transform–Load Technology". International Journal of Data Warehousing and Mining 5 (3): 27. doi:10.4018/jdwm.2009070101.
- ↑ Brown, S.A. (2016). "Patient Similarity: Emerging Concepts in Systems and Precision Medicine". Frontiers in Physiology 7: 561. doi:10.3389/fphys.2016.00561. PMC PMC5121278. PMID 27932992. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5121278.
- ↑ Aamodt, A.; Plaza, E. (1994). "Case-based reasoning: foundational issues, methodological variations, and system approaches". AI Communications 7 (1): 39–59.
- ↑ Motik, B.; Shearer, R.; Horrocks, I. (2009). "Hypertableau Reasoning for Description Logics". Journal Of Artificial Intelligence Research 36: 165–228. doi:10.1613/jair.2811.
- ↑ Singhal, A.; Simmons, M.; Lu, Z. (2016). "Text Mining Genotype-Phenotype Relationships from Biomedical Literature for Database Curation and Precision Medicine". PLoS Computational Biology 12 (11): e1005017. doi:10.1371/journal.pcbi.1005017. PMC PMC5130168. PMID 27902695. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5130168.
- ↑ Gene Ontology Consortium (2008). "The Gene Ontology project in 2008". Nucleic Acids Research 36 (DB1): D440-4. doi:10.1093/nar/gkm883. PMC PMC2238979. PMID 17984083. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2238979.
- ↑ Ghosh, S.; Matsuoka, Y.; Asai, Y. et al. (2011). "Software for systems biology: From tools to integrated platforms". Nature Reviews Genetics 12 (12): 821-32. doi:10.1038/nrg3096. PMID 22048662.
- ↑ Hucka, M.; Bergmann, F.T.; Hoops, S. et al. (2015). "The Systems Biology Markup Language (SBML): Language Specification for Level 3 Version 1 Core". Journal of Integrative Bioinformatics 12 (2): 266. doi:10.2390/biecoll-jib-2015-266. PMC PMC5451324. PMID 26528564. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5451324.
- ↑ Ganslandt, T.; Mate, S.; Helbing, K. et al. (2011). "Unlocking Data for Clinical Research - The German i2b2 Experience". Applied Clinical Informatics 2 (1): 116–27. doi:10.4338/ACI-2010-09-CR-0051. PMC PMC3631913. PMID 23616864. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3631913.
- ↑ Firnkorn, D.; Merker, S.; Ganzinger, M. et al. (2016). "Unlocking Data for Statistical Analyses and Data Mining: Generic Case Extraction of Clinical Items from i2b2 and tranSMART". Studies in Health Technology and Informatics 228: 567–71. PMID 27577447.
- ↑ Bach, K.; Sauer, C.; Althoff, K.-D.; Roth-Berghofer, T. (2014). "Knowledge Modeling with the Open Source Tool myCBR". Proceedings of the 10th Workshop on Knowledge Engineering and Software Engineering 2014. https://sds.dfki.de/publication/knowledge-modeling-open-source-tool-mycbr.
Notes
This presentation is faithful to the original, with only a few minor changes to grammar, spelling, and presentation, including the addition of PMCID and DOI when they were missing from the original reference.