Chemometric analysis of cannabinoids: Chemotaxonomy and domestication syndrome
| Full article title | Chemometric analysis of cannabinoids: Chemotaxonomy and domestication syndrome |
|---|---|
| Journal | Scientific Reports |
| Author(s) | Mudge, E.M.; Murch, S.J.; Brown, P.N. |
| Author affiliation(s) | University of British Columbia, British Columbia Institute of Technology |
| Primary contact | Email: Send message through journal website |
| Year published | 2018 |
| Volume and issue | 8 |
| Page(s) | 13090 |
| DOI | 10.1038/s41598-018-31120-2 |
| ISSN | 2045-2322 |
| Distribution license | Creative Commons Attribution 4.0 International |
| Website | https://www.nature.com/articles/s41598-018-31120-2 |
| Download | https://www.nature.com/articles/s41598-018-31120-2.pdf (PDF) |
Abstract
Cannabis is an interesting domesticated crop with a long history of cultivation and use. Strains have been selected through informal breeding programs with undisclosed parentage and criteria. The term “strain” refers to minor morphological differences and grower branding rather than distinct cultivated varieties. We hypothesized that strains sold by different licensed producers are chemotaxonomically indistinguishable and that the commercial practice of identifying strains by the ratio of total Δ9-tetrahydrocannabinol (THC) and cannabidiol (CBD) is insufficient to account for the reported human health outcomes. We used targeted metabolomics to analyze 11 known cannabinoids and an untargeted metabolomics approach to identify 21 unknown cannabinoids. Five clusters of chemotaxonomically indistinguishable strains were identified from the 33 commercial products. Only three of the clusters produce cannabidiolic acid (CBDA) in significant quantities, while the other two clusters redirect metabolic resources toward the tetrahydrocannabinolic acid (THCA) production pathways. Six unknown metabolites were unique to CBD-rich strains and/or correlated to CBDA, and three unknowns were found only in THC-rich strains. Together, these data indicate the domestication of the Cannabis germplasm has resulted in a loss of the CBDA pathway in some strains and reallocation of resources between CBDA and THCA pathways in others. The impact of domestication is a lack of chemical diversity and loss of biodiversity in modern Cannabis strains.
Introduction
Cannabis sativa L. (marijuana) is a dioecious, annual plant from Central Asia that has been used medicinally and recreationally for thousands of years.[1] The domestication of Cannabis has included human selection, inbreeding, and cross breeding, as well as natural outcrossing and genome mixing.[1] Strains are not easily delineated by genotype, and only moderate correlations have been observed between C. indica and C. sativa ancestry. In addition, large genetic variance has been observed within identically named strains.[2][3] Standardized, highly controlled programs to breed elite varieties or cultivars by selection of phytochemical profile have been limited.[4][5] It is estimated that there are several hundred or perhaps thousands of strains of cannabis currently being cultivated in legal and illegal markets.[4] It is possible that chemically identical or very closely related plant material is being sold under several different names by different producers, with no clear definition of the concept of a “strain.”
Cannabis producers market their products based on the amounts of total THC and CBD with the assumption that the overall phytochemical composition of the material can be extrapolated from these values, but there is considerable anecdotal evidence suggesting that strains with similar THC/CBD content have different effects on human physiology.[6][7] More than 120 different cannabinoids have been described in Cannabis[8][9], with the most interesting phytochemistry found in the glandular trichomes on the flowers of the female inflorescences.[10] THC is the most researched cannabinoid, and there are 10 additional classes of cannabinoids with varying chemical structures.[8] Cannabinoids are synthesized in acidic forms through the condensation of geranyl diphosphate (GPP) and most commonly olivetolic acid, products of the methylerythritol phosphate (MEP) and polyketide pathways.[11][12] There are several other polyketides that can be used in place of olivetolic acid, which contribute to the wide variation within this chemical class.[13][14] Neutral cannabinoids are products of decarboxylation from processing and handling harvested flowers.
Chemometric models are used to evaluate metabolite datasets to delineate relationships and identify potential influences on phytochemical diversity.[15][16][17] These approaches can be classified as targeted analysis, untargeted phytochemical discovery, metabolomic profiling. or fingerprinting.[15] Targeted metabolomics determines differences in known phytochemicals, while the untargeted approaches evaluate unidentified compounds in the phytochemical profiles.[15] Targeted-untargeted approaches combine known metabolites with the untargeted datasets as a hypothesis-generating tool to discover metabolite relationships, clusters, families and biochemical pathways.[15][18] The use of these models and algorithms enables a better understanding of metabolite commonality and diversity within plant species.[19]
We hypothesized that the total THC and CBD content is not sufficient to distinguish strains and that a combination of targeted and untargeted chemometric approaches can be used to predict cannabinoid composition and to better understand the impact of informal breeding program and selection on the phytochemical diversity of Cannabis. To investigate these hypotheses, we assembled a collection of Cannabis strains sold by licensed producers in Canada primarily based on total THC/CBD content, and analyzed the strains for known cannabinoids using a previously validated analytical method[20] to establish clusters of similar plant materials.
We then used an untargeted metabolomics approach to identify previously uncharacterized compounds and chemical relationships. We identified five clusters of chemotaxonomically indistinguishable strains within the collection. Our results show that the variation in less abundant cannabinoids between Cannabis strains was not dependent on the total THC and CBD content. These data suggest that the domestication of the Cannabis germplasm has resulted in the loss of the CBDA pathway in some strains and the reallocation of resources between CBDA and THCA pathways in others.
Results
Targeted metabolomics of cannabinoids
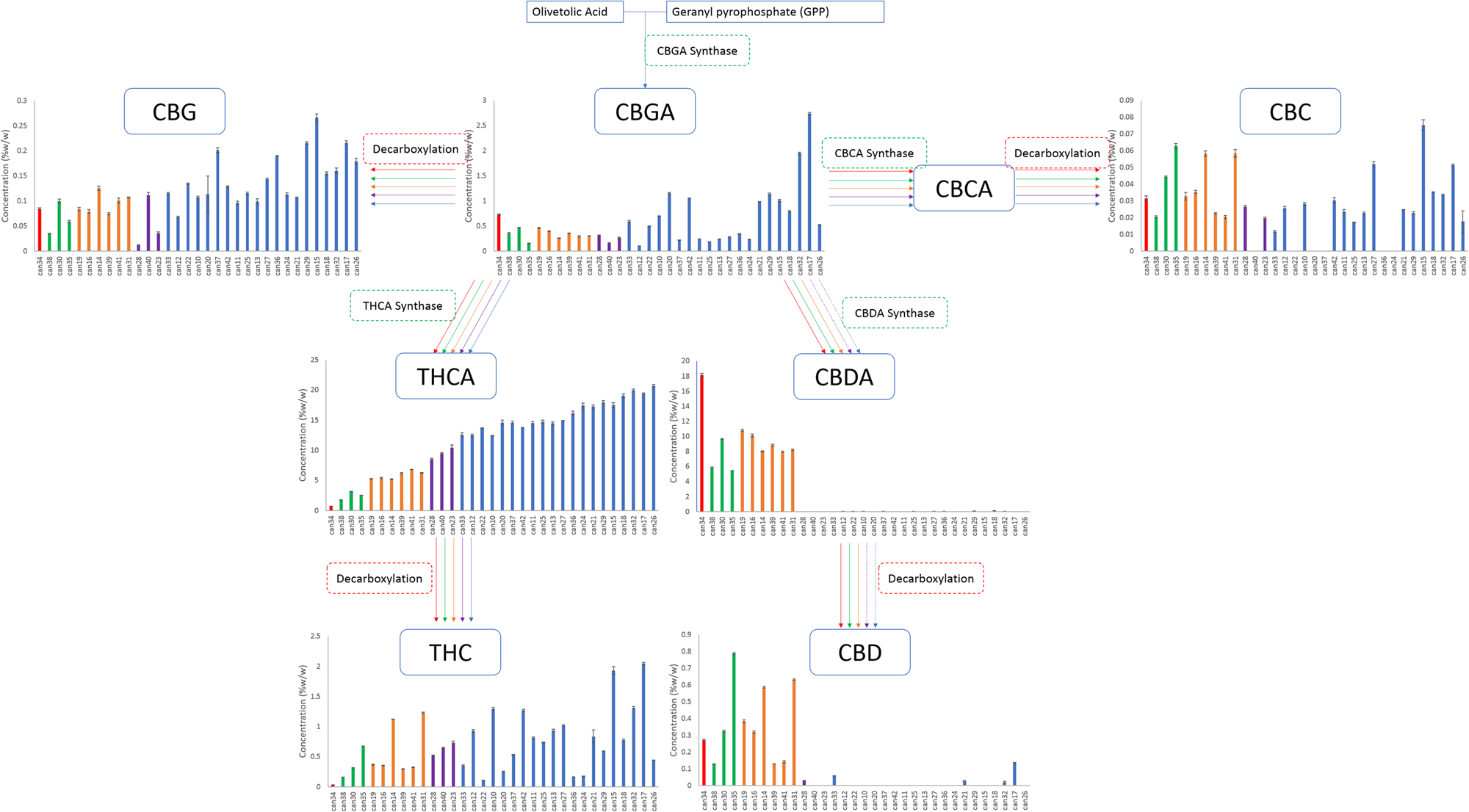
Two cannabinoids for which standards were obtained, cannabidivarin (CBDV) and cannabicyclol (CBL), were not detected in any strain. The 11 remaining cannabinoids with available chemical reference standards were identified and quantified. THCA content ranged from 0.76 to 20.71% w/w, with almost a linear increase in content from the lowest to highest strain with an r2 of 0.97, while CBDA content ranged from <MDL to 18.11% w/w, with the highest CBDA strains having the lowest THCA contents (Fig. 1). In THC-abundant strains, the CBDA levels were less than 0.15%, while in CBD abundant strains the content was greater than 5%. THC, the decarboxylated form of THCA, was present in strains from <LOQ up to 2% by weight in some strains, while CBD contents ranged from <MDL to 0.8%. CBD was most prevalent in high-CBDA strains. In addition, seven cannabinoids present at lower levels were quantified using individual calibration standards: tetrahydrocannabivarin (THCV), cannabigerol (CBG), cannabinol (CBN), cannabichromene (CBC), cannabidivarinic acid (CBDVA), cannabigerolic acid (CBGA), and Δ8-THC.
|
Classification of strains
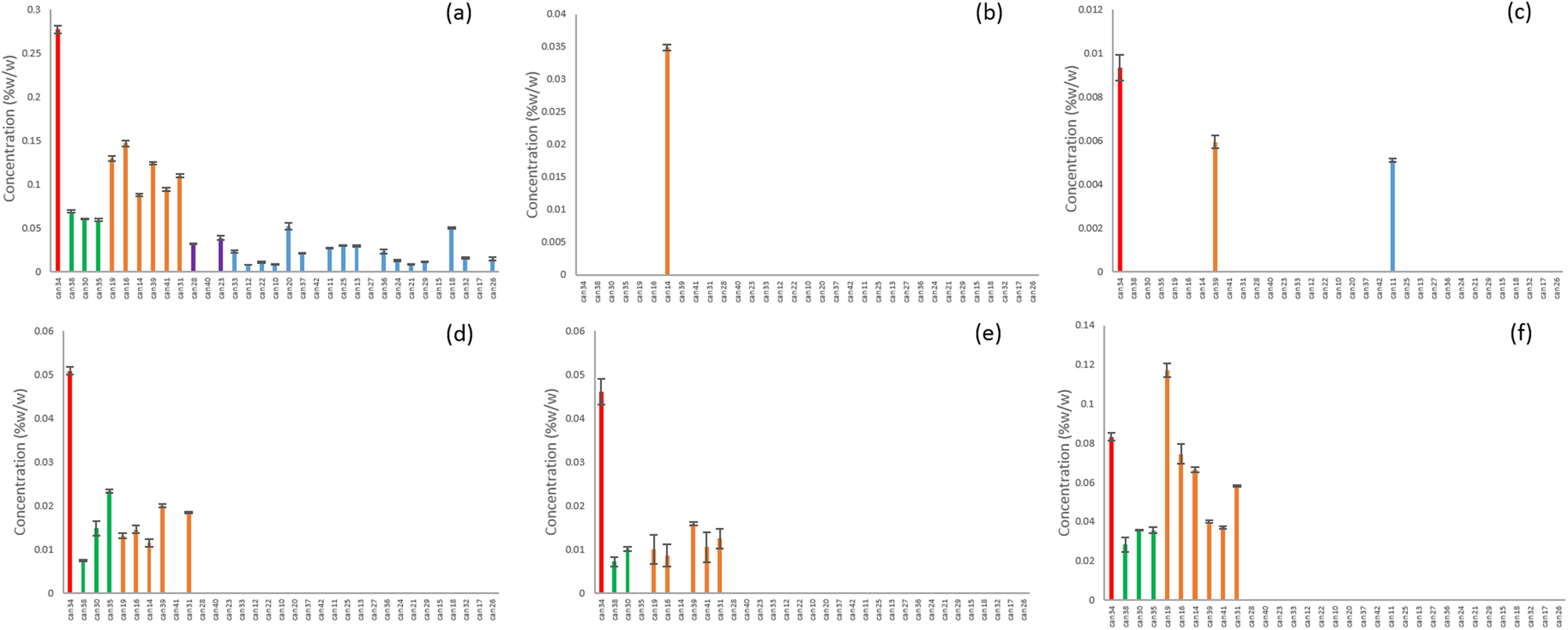
We hypothesized that individual plant breeders selected for Cannabis strains by up-regulating and down-regulating specific enzymes within the biosynthetic pathways, resulting in a redirection of metabolites between THCA and CBDA. Our data analysis identified five clusters of strains that fall within a narrow range of total CBD/THC values consistent with this hypothesis (Table 1). The branch of the biosynthetic pathway with olivetolic acid and geranyl pyrophosphate as precursors produces CBGA, CBG, CBCA, CBC, THCA, THC, CBDA and CBD (Fig. 1). Strains from all clusters contained measurable amounts of CBGA, CBG, THCA and THC (Fig. 1). Nine strains from the clusters with higher concentrations of THCA (blue and purple) did not contain detectable levels of CBC (Fig. 1). Two of the clusters were not found to contain significant quantities of CBDA and CBD (Fig. 1; blue and purple). One strain was different from all others and had a greater CBDA content and detectable levels of CBGA, CBG, CBC, and CBD with minimal THCA and THC (Fig. 1; red).
| |||||||||||||||||||||||||||||||||||
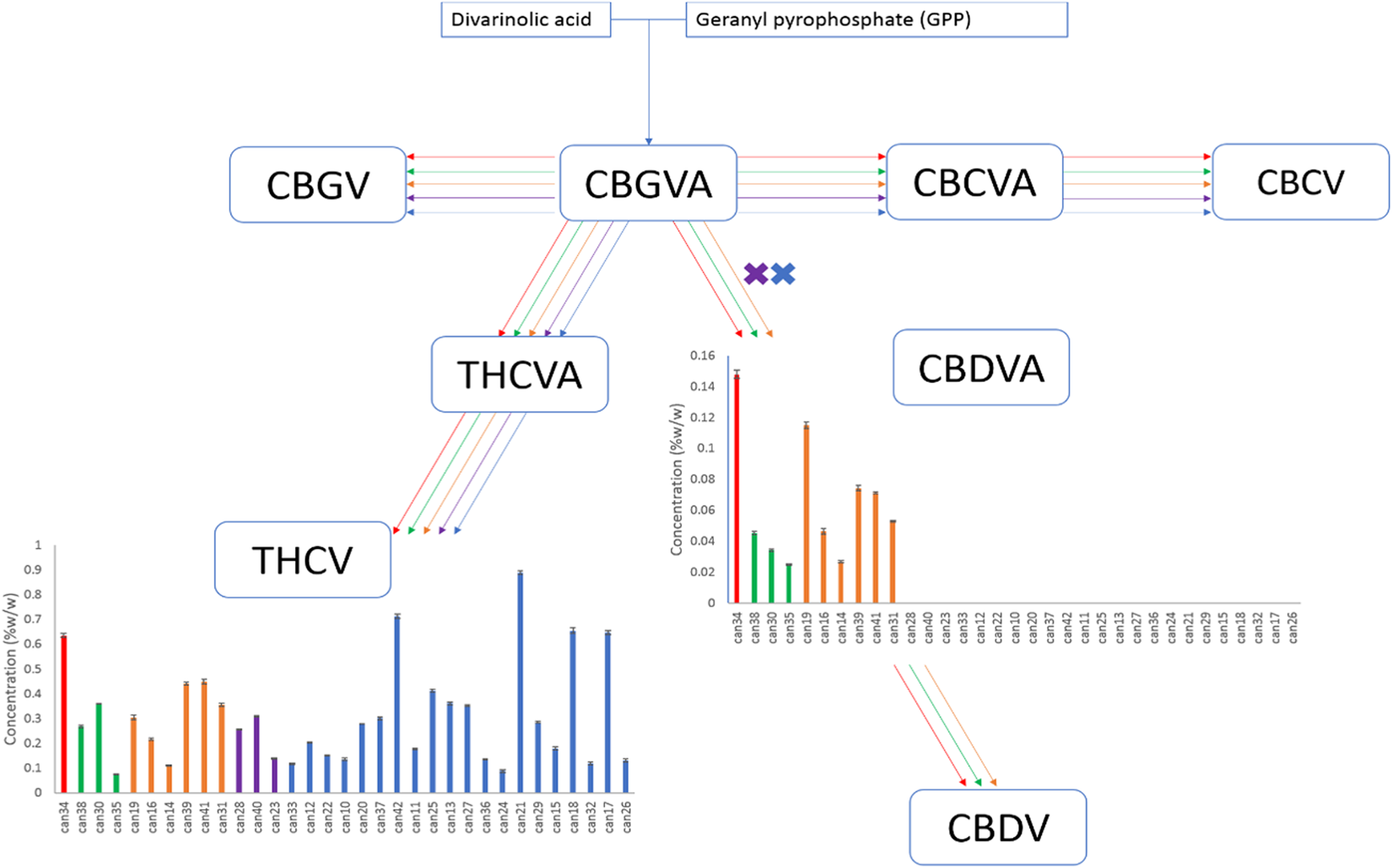
Compounds produced from the precursors divarinolic acid and geranyl pyrophosphate via cannabigerovarin acid (CBGVA) were also found to differ by strain cluster (Fig. 2). CBGVA appears to be a branch point for allocation of resources in Cannabis between THCV and CBDVA, indicating that the enzyme activity or the resource allocation mechanism for production of THCV was lost in the breeding process of strains clustered in the red, orange, and green groups (Fig. 2).
|
Untargeted metabolomics analysis
In addition to the 11 cannabinoids that corresponded with authentic standards, 21 peaks were identified in the chromatograms with UV spectra characteristic of cannabinoids. By comparison to THC, the contents were estimated from <MDL up to 0.34% by weight. Two unknown cannabinoids (CMPD-7 and CMPD-11) were detected in all strains, while CMPD-3 and CMPD-20 were each only detected in a single strain.
Relationships between known and unknown cannabinoids
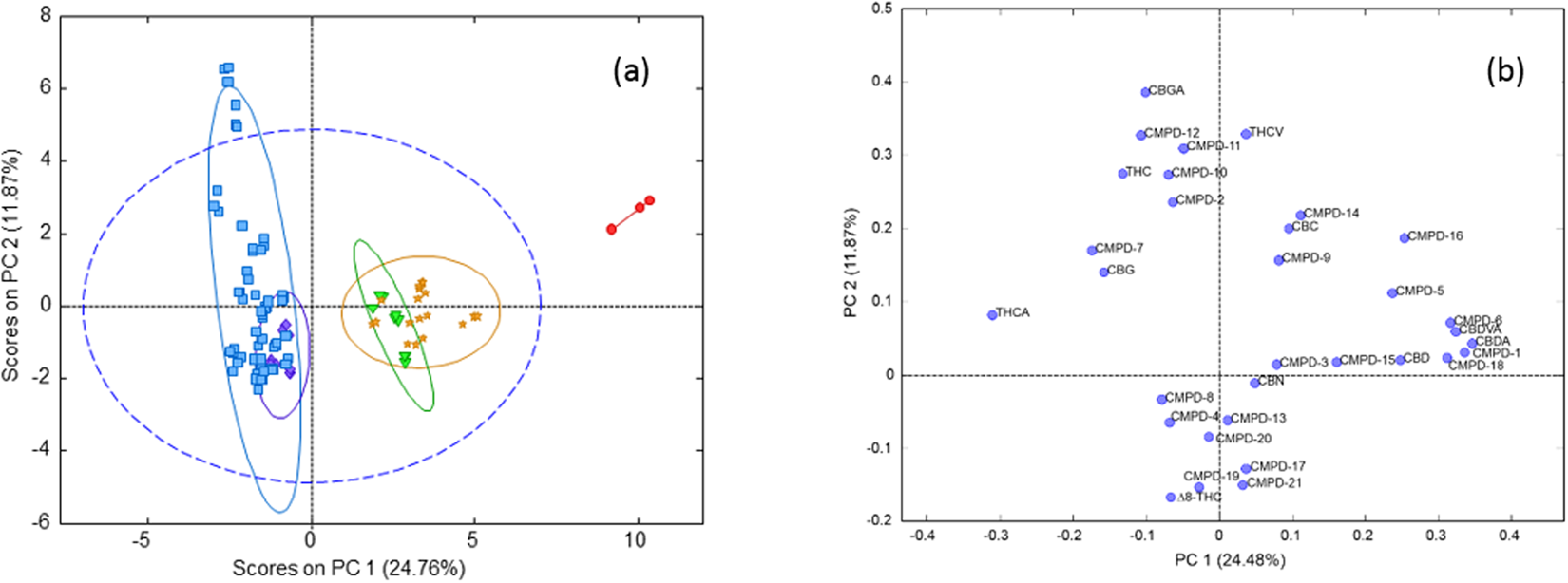
A principal component analysis (PCA) of the autoscaled cannabinoid data was plotted to show the clustering of the samples in an unsupervised fashion (Fig. 3). In the PCA plot, the first two principal components (PC) captured 36.6% of the variance in the data. Based on the loadings plot, the first PC was most highly influenced by the THCA and CBDA content of the strains, which are negatively correlated. There are two high THC strains (CAN17 and CAN21) and one CBD strain (CAN34) that were separated from the data clustered within the 95% confidence limit of the total data variance. Based on the loadings plot (Fig. 3B), CAN17 and CAN21 may be influenced by a significant number of low abundance cannabinoids including CBGA, CMPD-12, and CMPD-11. CAN34 is likely due to its significantly higher CBDA content relative to the other strains and because it contained less than 1% total THC.
|
While the first two principal components of PCA describe 36% of the variance, there is a remaining 64% of the variance in the cannabinoids not being described with this model. Therefore, additional models were employed to understand the relationships between cannabinoids and to identify additional strain classes based on the content of these 32 different cannabinoids. Multiple linear regression (MLR) analysis showed that 14 cannabinoids were better suited compared to all cannabinoids for predicting THCA content with validation r2 values improving from 0.02 and 0.88, respectively and for predicting CBDA content 14 cannabinoids improved the validation r2 values from 0.49 to 0.95 when compared with using the entire data set.
Pearson correlations were used to determine whether any of the unidentified cannabinoids could be associated with the major cannabinoids THCA, THC, CBDA, and CBD (Table 2). There was no significant correlation of THCA or THC and any of the unknown compounds (Table 2). The CBDA content was positively correlated with CMPD1, CBDVA, CMPD5, CMPD6, CMPD16, and CMPD18 (Table 2). CBD was potentially weakly correlated with CMPD1, CMPD6, and CBDA (Table 2).
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Putative identifications and pathways
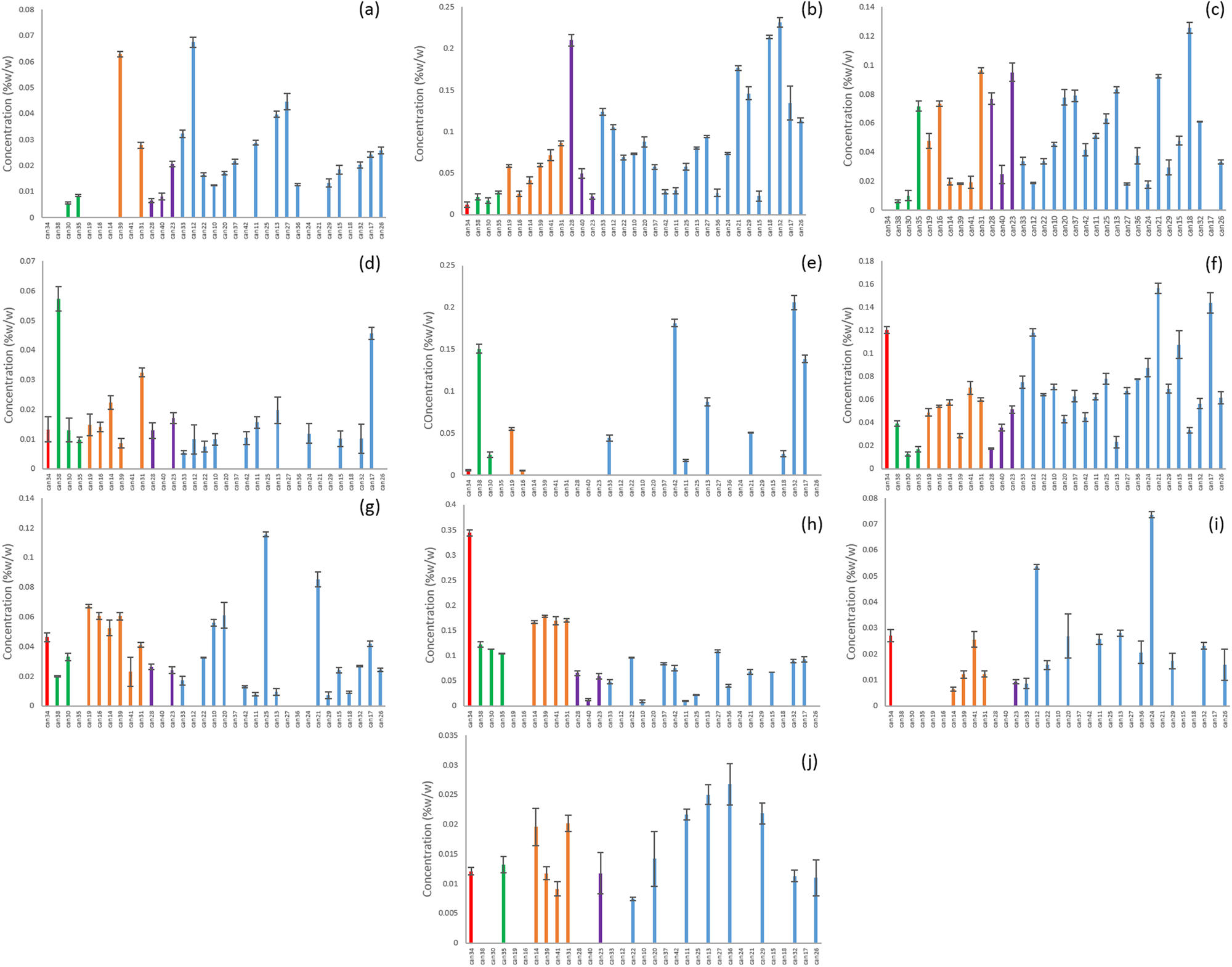
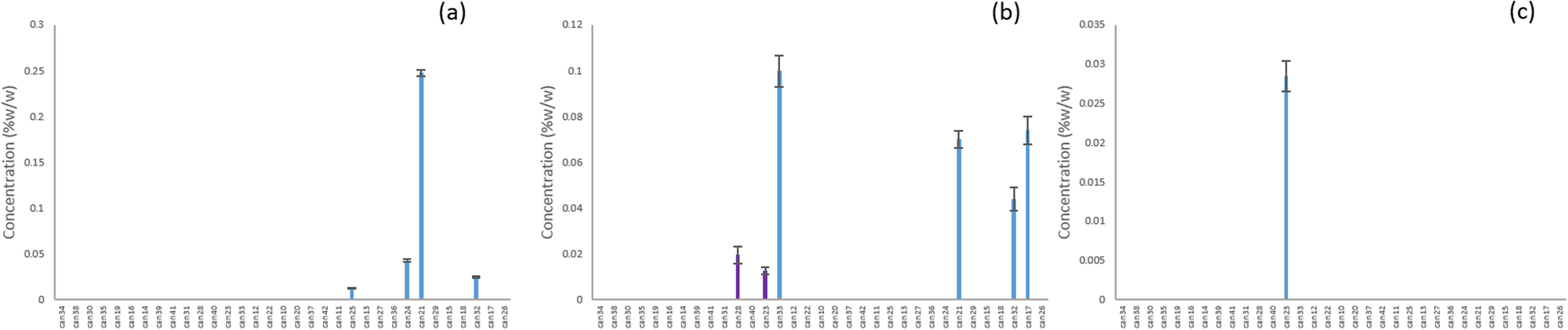
Ten of the unknowns were found across multiple strains from all of the clusters (Fig. 4). CMPD1 was strongly correlated with CBDA according to Pearson’s correlation (Table 2), and although it was found in many of the strains classified as blue or purple, it was at much higher concentrations in the red, green, and orange clusters (Fig. 5a). Compounds 3, 5, 6, 15, and 18 were found only in the CBD-rich clusters red, green, and orange (Fig. 5b–f). Compounds 2, 12, and 20 were found only in THC-dominant strains (Fig. 6a–c).
|
|
|
Discussion
The long history of human use has made the exact region of origin for Cannabis difficult to establish, though literature supports Northeast Asia.[1][4][21] Breeding of Cannabis cultivars in the hemp industry has focused on morphological improvements through established breeding programs, while marijuana, or drug-type cannabis, has primarily taken place in underground/clandestine programs through crossing landraces and/or indica and sativa lineages.[22][23] The major focus of breeding was increasing the yield of THC, although other features were considered, including organoleptics (aroma), morphology, color, and trichome density.[1][4][24] The genetic diversity between marijuana strains is lower in comparison with hemp varieties due to crossing closely related varieties.[2][3] CBDA and THCA synthases are thought to be controlled by two alleles on a single locus (B), crossing of CBDA and THCA dominant strains will produce offspring with intermediate total THC:CBD ratios.[5][25] With the prevalence of propagation through cuttings of mother plants, feminization of seeds, and production of sensimilla, the need for male plants has decreased, resulting in potential loss of genetic and phytochemical diversity.[1] The sativa and indica lineages used to describe Cannabis throughout the industry are based on postulation that sativa strains originated from European hemp cultivars, while indica are from potent, resinous Indian Cannabis[4], but given the use and trade of the plant in ancient times, the exact origin is unknown and these may not be distinct species.[21] Modern strains are considered dominant in either of these two “lineages” or hybrids between close relatives. These classifications focus on the pharmacological effects associated with the strains where sativa plants are considered stimulating and indica plants are associated with relaxation and sedation, but this is not a botanical or chemotaxonomical classification.[23] Comparisons of cannabinoid contents of these classifications have shown that the THC content can be identical between these two classification groups.[3][26] Many questions remain: What is a “strain”? Does a “strain” represent a phytochemically unique variety? Are “strains” from different growers actually different? Is there a more appropriate way to classify “strains”? Are these cultivars, varietals, landraces or even species? What is the impact of domestication on the ecological fitness of the species?
Breeding closely related plants potentially leads to loss of genetic diversity within the genome.[27] Traits signifying domestication syndrome include phenotypic changes such as increase seed size, loss of shattering, changes in reproduction, changes in secondary metabolites and loss of pest resistance compared with wild ancestors.[27][28] Reviews of Cannabis breeding have summarized domestication in terms of morphology, while focus on secondary metabolism has focused primarily on THC content.[1][4] Recent forensic evaluations of confiscated sensimilla Cannabis in the U.S. has shown dramatic increases in total THC content over the last 30 years, from 6.3% to 11.5%[29][30], but strains with greater than 20% total THC are available in the marketplace. This artificial increase in THCA production has resulted in the loss of CBDA synthase activity in THC dominant strains. Although crossbreeding will result in THC:CBD hybrid offspring, the loss of other biosynthetic pathways is unknown due to the non-rigorous breeding programs focusing primarily on the production of a single metabolite. Our data indicate that these breeding programs have also impacted unknown related metabolites with undetermined function.
Metabolomics analysis can generate chemotaxonomic classifications of plants in addition to hypothesis-generating insight of data correlations, metabolite identification, and relationships that would not be possible through single metabolite evaluation.[15][31] Using the correlation data and PCA loadings plots, we can hypothesize the putative identity of some of these unknown cannabinoids. For example, CMPD6 had a Pearson correlation coefficient of 0.89 with CBDA and occupies the same space within the PCA loadings plot. The UV spectra with a maximum of 224 nm identifies this compound as an acidic cannabinoid which was only detected in the presence of CBDA. Further evaluation showed that it eluted between CBDVA and CBDA; therefore, is hypothesized to be CBDA-C4 with a butyl side chain on the polyketide (Table 2).[32] Likewise, we putatively identified CBDA-C1, CBDM, and CBDMA among the unknowns separated by our chromatography protocol (Table 2). Due to the presence of THCA synthase in all strains, the correlation cannabinoids produced with this enzyme is less obvious. It was previously reported that low abundance cannabinoids may be regulated by upstream biosynthesis of precursor polyketides.[14] We found fewer unknown cannabinoids in the strains selected for higher THC content. With such strong emphasis on the synthesis of a single metabolite, there is a strong possibility that other biosynthetic pathways have been lost in the process.[27][28]
Several classification systems have been proposed for Cannabis based on a limited number of phenotypic attributes.[1][4][33] The concept of a “strain” does not reflect the crop domestication, breeding programs, or plant chemistry. The strains available in the Canadian marketplace are closely related, and evaluating single metabolite classes does not provide sufficient information to understand the phytochemical diversity available. The abundance of secondary metabolites within plants does not necessarily correlate with pharmacological significance, and with Cannabis there is the postulated “entourage effect” describing the synergistic effects of many metabolites for anecdotal medical efficacy.[7] Domestication of the crop has limited the genetic variability in the crop, and the impact on crop diversity, physiology, and metabolism is not fully understood. Further research is needed to evaluate the low abundance cannabinoids for potential medicinal efficacy and to determine their roles in plant metabolism.
Methods
Reagents
Methanol, acetonitrile, ammonium formate and formic acid (98%) were HPLC grade. Water was deionized and purified to 18.2 MΩ using a Barnstead Smart2Pure nanopure system (Thermo Scientific). Cannabinoid standards for quantification were purchased from Cerilliant Corp. (Round Rock, TX) for tetrahydrocannabinolic acid (THCA), THC, cannabidiolic acid (CBDA), CBD, cannabigerol (CBG), cannabichromene (CBC), tetrahydrocannabivarin (THCV) and cannabinol (CBN), Δ8-THC, cannabidivarinic acid (CBDVA), cannabidivarin (CBDV), cannabigerolic acid (CBGA) and cannabicyclol (CBL). All standards were provided as 1.0 mg/mL solutions in either methanol or acetonitrile.
Plant materials
Thirty-three strains of Cannabis were purchased from five licensed producers in Canada under the Access to Cannabis for Medical Purposes Regulations, and laboratory analysis was performed under a Health Canada Controlled Drugs and Substances License. The test samples were provided as whole or milled flowers in five, 10, and 15 gram packages and stored at room temperature until use. Due to the legal restrictions pertaining to the storage of Cannabis strains, submission of voucher specimens to a herbarium were not possible, but given the regulatory framework associated with these plants, their identify has been confirmed as Cannabis sativa L.
Targeted metabolomics of cannabinoids
The content of 13 cannabinoids was determined according to a previously validated analytical method.[20] In brief, ground Cannabis flowers (0.200 g) were extracted with 25 mL of 80% methanol in a 50 mL amber centrifuge tube for 15 minutes by sonication at room temperature with vortexing every five minutes, followed by centrifugation at 4500 g for five minutes and filtration with a 0.22 µm PTFE filter. Extracts were diluted to within the calibration range using the extraction solvent and placed in the 4°C sample holder for same-day analysis. Chromatographic separation was performed on an Agilent 1200 UHPLC with a Kinetex C18 100 mm × 3.0 mm, 1.8 µm column (Phenomenex; Torrance, CA) using gradient elution with 10 mM ammonium formate (pH 3.6) and acetonitrile. The autosampler was maintained at 4°C and detection was at 220 nm. The peak areas for peaks with typical acidic or neutral cannabinoid UV spectra eluting between 2.5 and 14.5 minutes were collected using Chemstation software (Agilent Technologies), and known cannabinoids were identified. Known cannabinoids were quantified in % w/w against their individual calibration curves using external calibration in Microsoft Excel. The total THC content was determined as the sum of THC and THCA in addition to the total degradation products of THC: CBN and Δ8-THC, adjusted by the molar mass ratios. Total CBD content was determined as the sum of CBDA and CBD adjusted by molar mass ratios.
Untargeted metabolomics
Unknown cannabinoids were identified and numbered in sequential order as they appeared in the chromatogram. Unknown cannabinoids were quantified as THC equivalents using the THC calibration curves and ordered in sequential order in the chromatogram as CMPD#.
Data analysis
For multivariate analysis, missing values were replaced with the method detection limit (MDL) divided by two for each assigned cannabinoid. In the cannabinoid profiles, where the MDL has not been determined for unassigned peaks, the missing data was replaced with half of the MDL of THC. Pearson correlation coefficients to determine relationships between metabolites were calculated using the cor script in R. As the concentration of a given metabolite does not necessarily correlate with pharmacological activity, the data were autoscaled by mean centering and scaling to unit variance in order to give each metabolite equal weight prior to multivariate analyses. Principal component analysis (PCA) and multiple linear regression (MLR) analysis were subsequently modeled using Solo + MIA (Eigenvector Research).
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Acknowledgements
This research was undertaken, in part, thanks to funding from the Canada Research Chairs program.
Author contributions
E.M., S.M. and P.B. designed the study. E.M. conducted the laboratory analysis. All authors analyzed the data and wrote the manuscript. All authors gave final approval for publication.
Competiting interests
The authors declare no competing interests.
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Clarke, R.C.; Merlin, M.D. (2016). "Cannabis Domestication, Breeding History, Present-day Genetic Diversity, and Future Prospects". Critical Reviews in Plant Sciences 35 (5–6): 293–327. doi:10.1080/07352689.2016.1267498.
- ↑ 2.0 2.1 Sawler, J.; Stout, J.M.; Fardner, K.M. et al. (2015). "The Genetic Structure of Marijuana and Hemp". PLoS One 10 (8): e0133292. doi:10.1371/journal.pone.0133292. PMC PMC4550350. PMID 26308334. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4550350.
- ↑ 3.0 3.1 3.2 Soler, S.; Gramazio, P.; Figàs, M.R. et al. (2017). "Genetic structure of Cannabis sativa var. indica cultivars based on genomic SSR (gSSR) markers: Implications for breeding and germplasm management". Industrial Crops and Products 104: 171–78. doi:10.1016/j.indcrop.2017.04.043.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 Small, E. (2015). "Evolution and Classification of Cannabis sativa (Marijuana, Hemp) in Relation to Human Utilization". The Botanical Review 81 (3): 189–294. doi:10.1007/s12229-015-9157-3.
- ↑ 5.0 5.1 de Meijer, E. (2014). "Chapter 5: The Chemical Phenotypes (Chemotypes) of Cannabis". In Pertwee, R.. Handbook of Cannabis. Oxford Scholarship Online. pp. 89–110. doi:10.1093/acprof:oso/9780199662685.003.0005. ISBN 9780199662685.
- ↑ McPartland, J.M.; Russo, E.B. (2001). "Cannabis and Cannabis Extracts". Journal of Cannabis Therapeutics 1 (3–4): 103–32. doi:10.1300/J175v01n03_08.
- ↑ 7.0 7.1 Russo, E.B. (2011). "Taming THC: potential cannabis synergy and phytocannabinoid-terpenoid entourage effects". British Journal of Pharmacology 163 (7): 1344–64. doi:10.1111/j.1476-5381.2011.01238.x. PMC PMC3165946. PMID 21749363. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3165946.
- ↑ 8.0 8.1 ElSohly, M.a.; Gul, W. (2014). "Chapter 1: Constituents of Cannabis sativa". In Pertwee, R.. Handbook of Cannabis. Oxford Scholarship Online. pp. 3–22. doi:10.1093/acprof:oso/9780199662685.003.0001. ISBN 9780199662685.
- ↑ Turner, C.E.; ElSohly, M.A.; Boeren, E.G. (1980). "Constituents of Cannabis sativa L. XVII. A review of the natural constituents". Journal of Natural Products 43 (2): 169-234. PMID 6991645.
- ↑ Turner, J.C.; Hemphill, J.K.; Mahlberg, P.G. (1978). "Quantitative determination of cannabinoids in individual glandular trichomes of Cannabis sativa L. (Cannabacaea)". American Journal of Botany 65 (10): 1103–06. doi:10.1002/j.1537-2197.1978.tb06177.x.
- ↑ Flores-Sanchez, I.J.; Verpoorte, R. (2008). "Secondary metabolism in cannabis". Phytochemistry Reviews 7 (3): 615–639. doi:10.1007/s11101-008-9094-4.
- ↑ Fellermeier, M.; Eisenreich, W.; Bacher, A. et al. (2001). "Biosynthesis of cannabinoids. Incorporation experiments with (13)C-labeled glucoses". European Journal of Biochemistry 268 (6): 1596-604. PMID 11248677.
- ↑ Degenhardt, F.; Stehle, F.; Kayser, O. (2017). "Chapter 2: The Biosynthesis of Cannabinoids". In Preedy, V.R.. Handbook of Cannabis and Related Pathologies. Elsevier. pp. 13–23. doi:10.1016/B978-0-12-800756-3.00002-8. ISBN 9780128007563.
- ↑ 14.0 14.1 Shoyama, Y.; Hirano, H.; Nishioka, I. (1984). "Biosynthesis of propyl cannabinoid acid and its biosynthetic relationship with pentyl and methyl cannabinoid acids". Phytochemistry 23 (9): 1909–12. doi:10.1016/S0031-9422(00)84939-0.
- ↑ 15.0 15.1 15.2 15.3 15.4 Turi, C.E.; Finley, J.; Shipley, P.R. et al. (2015). "Metabolomics for phytochemical discovery: development of statistical approaches using a cranberry model system". Journal of Natural Products 78 (4): 953-66. doi:10.1021/np500667z. PMID 25751407.
- ↑ Worley, B.; Powers, R. (2013). "Multivariate Analysis in Metabolomics". Current Metabolomics 1 (1): 92–107. doi:10.2174/2213235X11301010092. PMC PMC4465187. PMID 26078916. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4465187.
- ↑ Hagel, J.M.; Facchini, P.J. (2008). "Plant metabolomics: analytical platforms and integration with functional genomics". Phytochemistry Reviews 7 (3): 479–497. doi:10.1007/s11101-007-9086-9.
- ↑ Brown, P.N;. Murch, S.J.; Shipley, P. (2012). "Phytochemical diversity of cranberry (Vaccinium macrocarpon Aiton) cultivars by anthocyanin determination and metabolomic profiling with chemometric analysis". Journal of Agricultural and Food Chemistry 60 (1): 261–71. doi:10.1021/jf2033335. PMID 22148867.
- ↑ Scherling, C.; Roscher, C.; Giavalisco, P. et al. (2010). "Metabolomics unravel contrasting effects of biodiversity on the performance of individual plant species". PLoS One 5 (9): e12569. doi:10.1371/journal.pone.0012569. PMC PMC2935349. PMID 20830202. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2935349.
- ↑ 20.0 20.1 Mudge, E.M.; Murch, S.J.; Brown, P.N. (2017). "Leaner and greener analysis of cannabinoids". Analytical and Bioanalytical Chemistry 409 (12): 3153–63. doi:10.1007/s00216-017-0256-3. PMC PMC5395585. PMID 28233028. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5395585.
- ↑ 21.0 21.1 Grof, C.P.L. (2018). "Cannabis, from plant to pill". British Journal of Clinical Pharmacology 84 (11): 2463–67. doi:10.1111/bcp.13618.
- ↑ Andre, C.M.; Hausman, J.F.; Guerriero, G. (2016). "Cannabis sativa: The Plant of the Thousand and One Molecules". Frontiers in Plant Science 7: 19. doi:10.3389/fpls.2016.00019. PMC PMC4740396. PMID 26870049. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4740396.
- ↑ 23.0 23.1 Piomelli, D.; Russo, E.B. (2016). "The Cannabis sativa Versus Cannabis indica Debate: An Interview with Ethan Russo, MD". Cannabis and Cannabinoid Research 1 (1): 44–6. doi:10.1089/can.2015.29003.ebr. PMC PMC5576603. PMID 28861479. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5576603.
- ↑ Sexton, M.; Shelton, K.; Haley, P. et al. (2018). "Evaluation of Cannabinoid and Terpenoid Content: Cannabis Flower Compared to Supercritical CO2 Concentrate". Planta Medica 84 (4): 234-241. doi:10.1055/s-0043-119361. PMID 28926863.
- ↑ de Meijer, E.P.; Bagatta, M.; Carboni, A. et al. (2003). "The inheritance of chemical phenotype in Cannabis sativa L.". Genetics 163 (1): 335–46. PMC PMC1462421. PMID 12586720. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1462421.
- ↑ Hazekamp, A.; Fischedick, J.T. (2012). "Cannabis - From cultivar to chemovar". Drug testing and analysis 4 (7–8): 660–7. doi:10.1002/dta.407. PMID 22362625.
- ↑ 27.0 27.1 27.2 Meyer, R.S.; DuVal, A.E.; Jensen, H.R. (2012). "Patterns and processes in crop domestication: An historical review and quantitative analysis of 203 global food crops". New Phytologist 196 (1): 29–48. doi:10.1111/j.1469-8137.2012.04253.x.
- ↑ 28.0 28.1 McKey, D.; Elias, M.; Pujol, B. et al. (2010). "The evolutionary ecology of clonally propagated domesticated plants". New Phytologist 186 (2): 318–32. doi:10.1111/j.1469-8137.2010.03210.x. PMID 20202131.
- ↑ ElSohly, M.A.; Ross, S.A.; Mehmedic, Z. et al. (2000). "Potency trends of delta9-THC and other cannabinoids in confiscated marijuana from 1980-1997". Journal of Forensic Sciences 45 (1): 24–30. PMID 10641915.
- ↑ Mehmedic, Z.; Chandra, S.; Slade, D. et al. (2010). "Potency trends of Δ9-THC and other cannabinoids in confiscated cannabis preparations from 1993 to 2008". Journal of Forensic Sciences 55 (5): 1209–17. doi:10.1111/j.1556-4029.2010.01441.x. PMID 20487147.
- ↑ Fiehn, O. (2002). "Metabolomics--The link between genotypes and phenotypes". Plant Molecular Biology 48 (1–2): 155–71. PMID 11860207.
- ↑ Smith, R.M. (1997). "Identification of Butyl Cannabinoids in Marijuanas". Journal of Forensic Sciences 42 (4): 610–18. doi:10.1520/JFS14173J.
- ↑ Small, E.; Beckstead, H.D. (1973). "Letter: Cannabinoid phenotypes in Cannabis sativa". Nature 245 (5421): 147–8. PMID 4582664.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation. Some grammar and punctuation was cleaned up to improve readability. In some cases important information was missing from the references, and that information was added.