Posted on June 17, 2024
By LabLynx
Journal articles
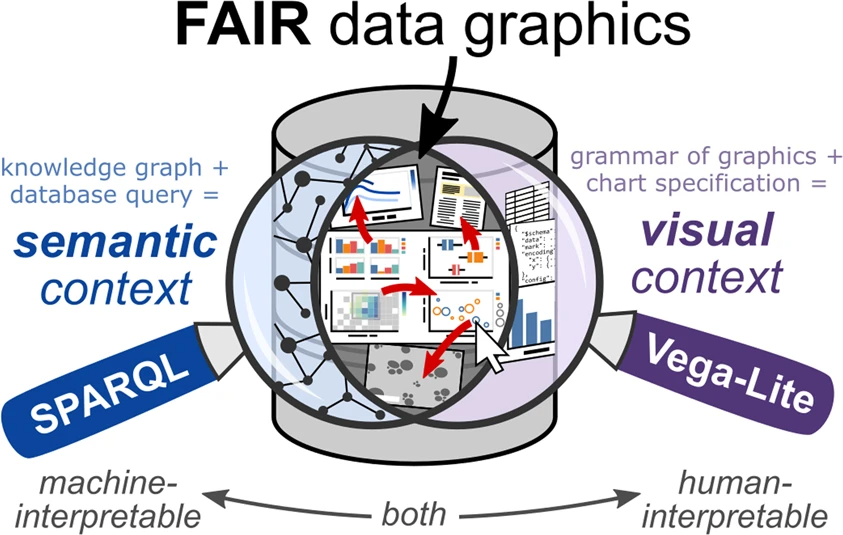
In this 2022 paper published in the journal
Scientific Data, Deagen
et al. present their approach to "web-based, interactive data graphics synchronized to a knowledge graph" in order to make materials science research objects more FAIR (findable, accessible, interoperable, and reusable). After a brief introduction, the authors discuss their methodology and results towards representing charts as metadata, tapping into SPARQL, Vega-Lite, and knowledge graph databases. They also address how their method touches upon multiple aspects of the FAIR data principles. They conclude that "altogether, this pairing of SPARQL and Vega-Lite—demonstrated here in the domain of polymer nanocomposite materials science—offers an extensible approach to FAIR ... scientific data visualization within a knowledge graph framework," adding that "defining charts as metadata in a knowledge graph captures semantic context and visual context while providing interactive, human-interpretable documentation of the contents of a knowledge graph."
Posted on June 11, 2024
By LabLynx
Journal articles
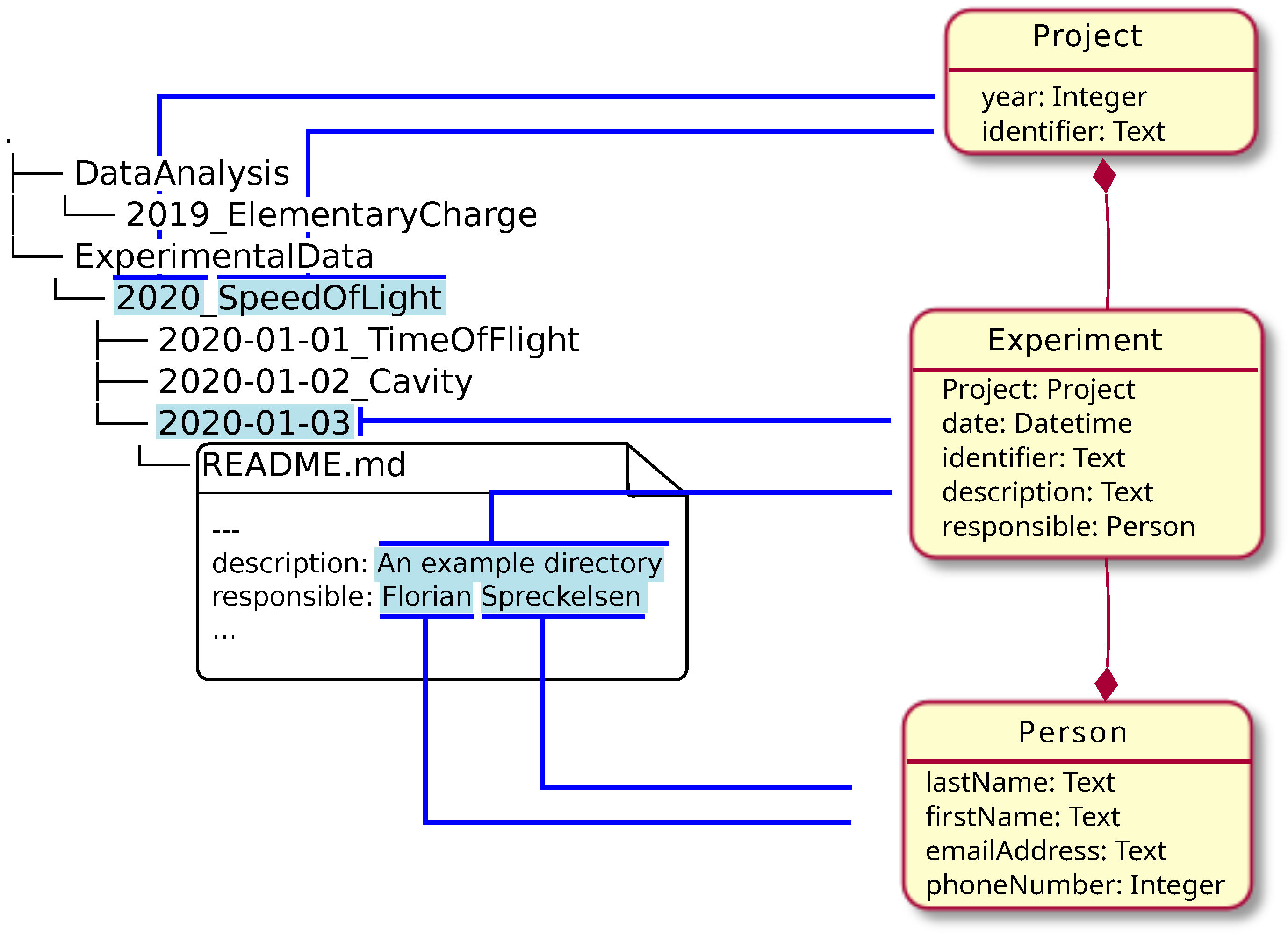
In this 2024 article published in the journal
Data, tom Wörden
et al. describe their approach to simultaneously using relational database systems with with more semantically driven research data management systems (RDMS) while largely keeping the two in synch. The authors use the open-source RDMS LinkAhead and its associated crawler, as well as YAML for synchronization and data mapping. They describe their approach in detail, noting the importance of "identifiables," which they define as "a set of properties that is taken from the file system layout or the file contents, which allow for uniquely identifying the corresponding object in the RDMS." After a brief discussion of their results and the limitations of their approach, they conclude that the practicality of their approach has been demonstrated, with the potential to transfer "the concept to multiple different use cases in order to make the crawler more robust" across a broader spectrum of scientific research settings.
Posted on June 4, 2024
By LabLynx
Journal articles
As the ramifications of the FAIR data principles of Wilkinson
et al. continue to shape how academic (and industrial) research is performed, it's inevitable that the information management systems used in these research contexts will be scrutinized under those same FAIR data principles. In this case, Plass
et al. of FAU Erlangen-Nürnberg describe how they landed upon the open-source electronic laboratory notebook (ELN) and laboratory information management system (LIMS) solution openBIS to address FAIR data management in their multidisciplinary Collaborative Research Centre (CRC). After discussing the current state of research data management (RDM), the authors go into the specifics of their implementation of openBIS, the adjustments they made to the system to address their information management needs, and the experiences they had using the modified system. They ultimately conclude that "this ELN-LIMS could serve as a template solution for other similarly structured collaborative research centers or research groups" given the benefits they saw with their modified version.
Posted on May 26, 2024
By LabLynx
Journal articles
In this "in press" journal article published in
Practical Laboratory Medicine, Smith
et al. of ARUP Laboratories provide the results of a survey conducted just prior to the U.S. Food and Drug Administration's (FDA's) April 29 release of their final rule on laboratory-developed tests (LDTs). Polling a select group of its clinical laboratory customers, the authors sought to discover clinical laboratorians' opinions about the then proposed changes to regulate LDTs as medical devices. The survey was conducted in part due to a set of perceived FDA shortcomings that would make it "difficult for the FDA to conduct an accurate quantitative analysis of public opinion in a key sector directly impacted by its proposed rule." The authors describe the structure of their survey and its associated analytical methods before discussing the results. They then provides a full discussion with a brief conclusion: "The majority of clinical laboratorians surveyed do not support the FDA’s proposed rule on LDTs and report having insufficient resources to comply with the rule if it is enacted," and that the FDA had more work to do "to better understand the negative impacts of the proposed rule."
Posted on May 21, 2024
By LabLynx
Journal articles
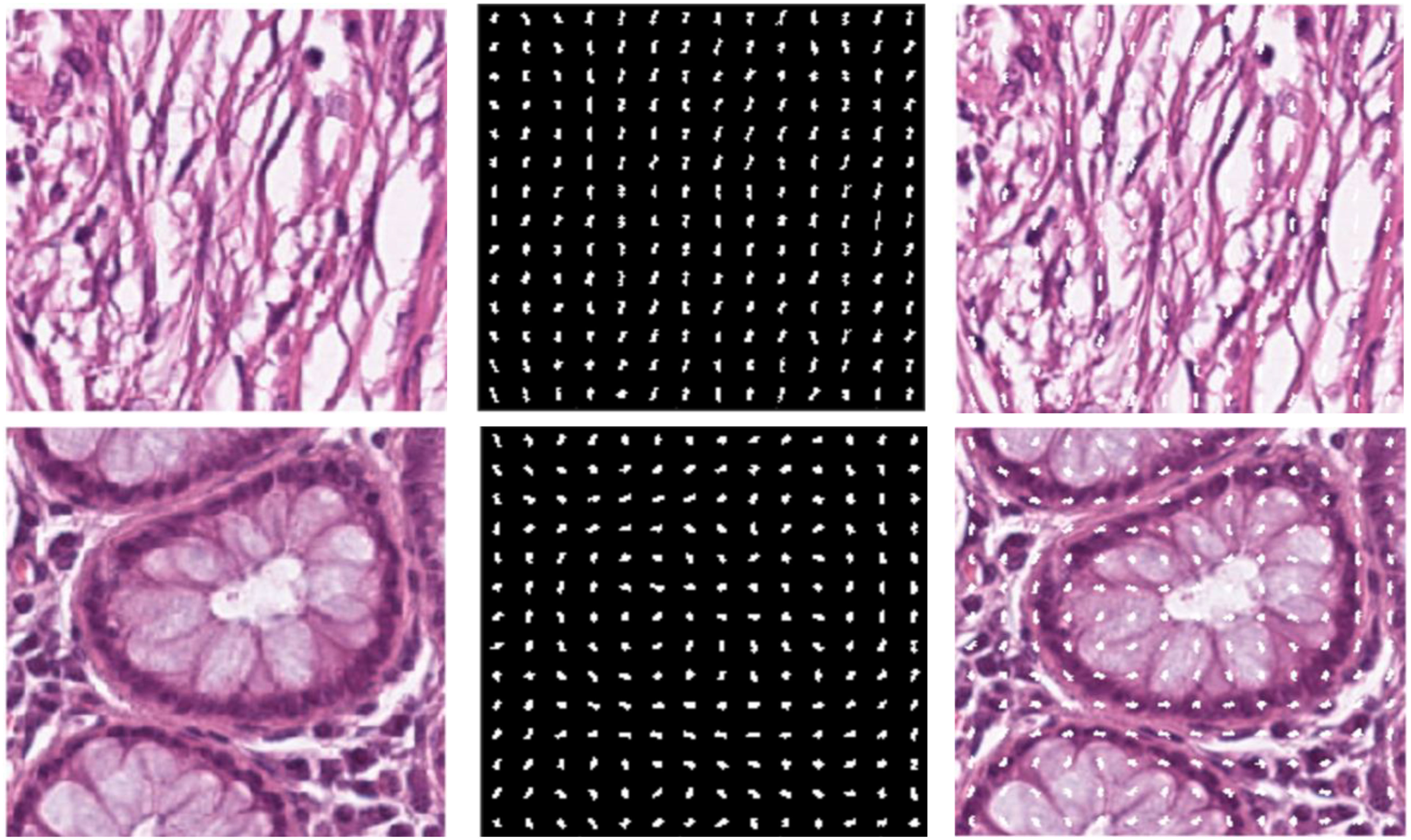
Histopathology diagnostics has largely been a manual process, with humans examining images of tissues and evaluating them based on knowledge passed down and experience gained. With the age of digital pathology reaching strong realization, new methods are arising, including with the use of artificial intelligence (AI). In this 2024 paper published in
Frontiers in Oncology, Doğan and Yılmaz present their AI-based system for image analysis, which taps into both a histogram of oriented gradients (HoG) method and a customized convolutional neural network (CNN) model. After discussing their approach and results, the authors discuss the implications of their results, concluding that their approach could be "considered a good alternative as it performs as well as and better than the more complex models described in the literature." They also say that their results highlight "the indisputable advantages of AI automation and the critical importance of data normalization and interpretation" for any AI-driven effort towards histopathology image analysis, while at the same time noting that the AI's "algorithms must [still] transparently reveal decision-making processes and be understandable by clinical experts."
Posted on May 13, 2024
By LabLynx
Journal articles
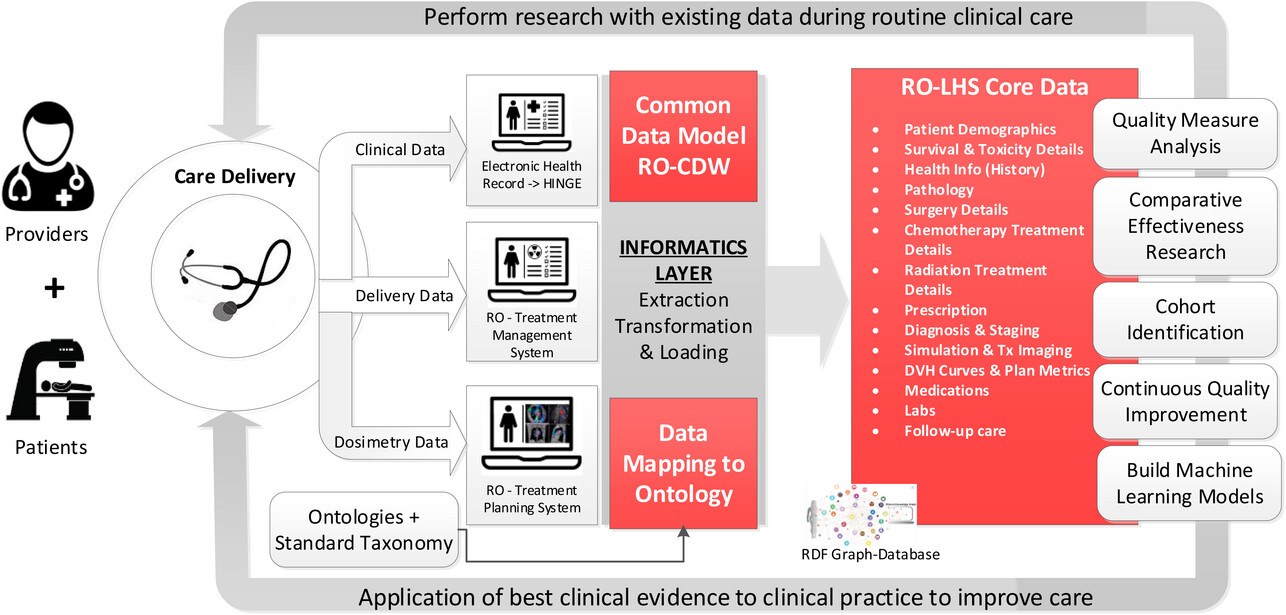
In this 2023 article published in the
Journal of Applied Clinical Medical Physics, Kapoor
et al. of Virginia Commonwealth University present their approach to managing radiation oncology research data in a FAIR (findable, accessible, interoperable, and reusable) way. The authors focus on the advantages of a knowledge graph-based database combined with ontology-based keyword search, synonym-based term matching, and a variety of visualization tools. After presenting background on the topic, Kapoor
et al. describe their approach using 1,660 patient clinical and dosimetry records, standardize terminologies, data dictionaries, and Semantic Web technologies. They then discussed the results of their mapping efforts, as well as the discovered benefits of a knowledge graph-based solution over a traditional relational database approach. They conclude that their approach "successfully demonstrates the procedures of gathering data from multiple clinical systems and using ontology-based data integration," with the radiation oncology datasets being more FAIR "using open semantic ontology-based formats," which in turn helps "facilitate interoperability and execution of large scientific studies."
Posted on May 7, 2024
By LabLynx
Journal articles
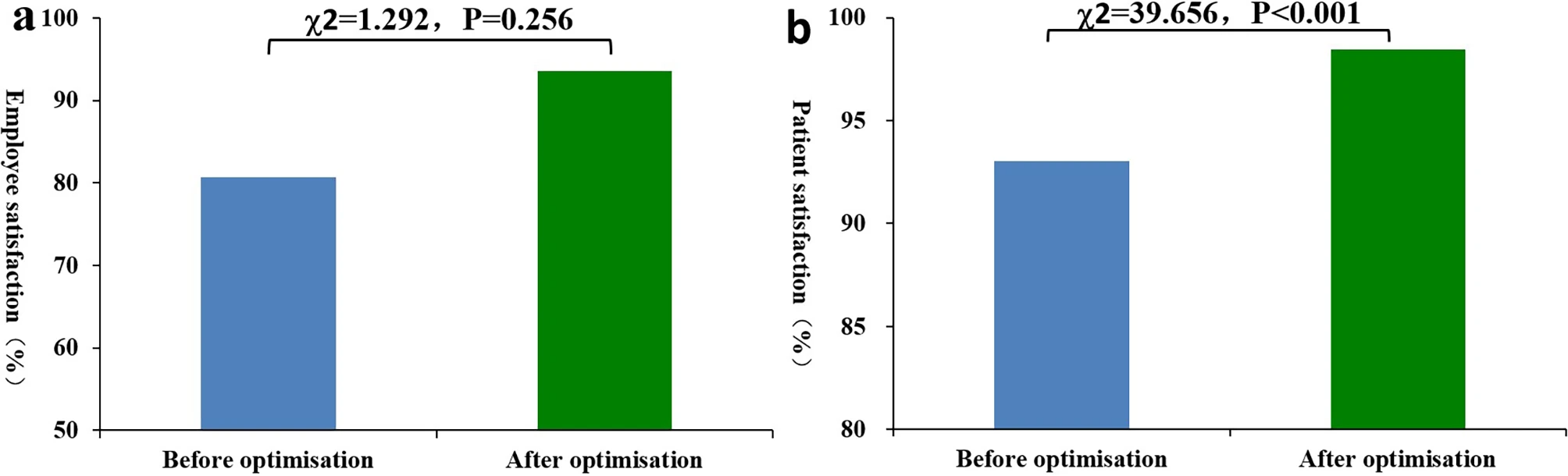
Can the thoughtful application and optimization of laboratory informatics solutions lead to do a variety of improvements in a hospital-based emergency medicine laboratory? Zhang
et al. sought to answer this question in a a contemporary tertiary second-class general hospital in China. After analyzing their emergency laboratory workflows, identifying problems withing those workflows, and identifying other areas of optimization, the authors applied a laboratory information management system (LIMS) to their operations, upgraded instrumentation, and finessed other aspects of their IT infrastructure. After discussing their results, the authors concluded that "the automation and standardization of most stages in emergency laboratory testing can be realized by IT, which reduces the workload of employees and improves emergency laboratory test quality," further noting that "emergency laboratory test report time can be shortened, emergency laboratory test quality can be enhanced, and employee and patient satisfaction can also be improved."
Posted on April 29, 2024
By LabLynx
Journal articles
In this brief article in
Journal of Privacy and Confidentiality, Jang
et al. of Inter-university Consortium for Political and Social Research (ICPSR) at University of Michigan review the current approach of researchers towards restricted data management and how it may need to evolve given certain challenges. They offer this review in the scope of increasing demands for FAIR (findable, accessible, interoperable, and reusable) data in research and journal communities. After a brief introduction, the authors discuss three main elements of managing restricted date: data use agreements, disclose review, and training. While the Five Safes framework is effective under this scope, the authors concludes that such "safeguards could generate unintended challenges to certain groups of individuals (e.g., institutional approval that could exclude researchers without institutional affiliation) or in different areas (e.g., rigorous output checking that requires extensive insights from experts)." They add that organizations may need develop more thoughtful and standardized data management policies that still remain flexible under the scope of FAIR data.
Posted on April 23, 2024
By LabLynx
Journal articles
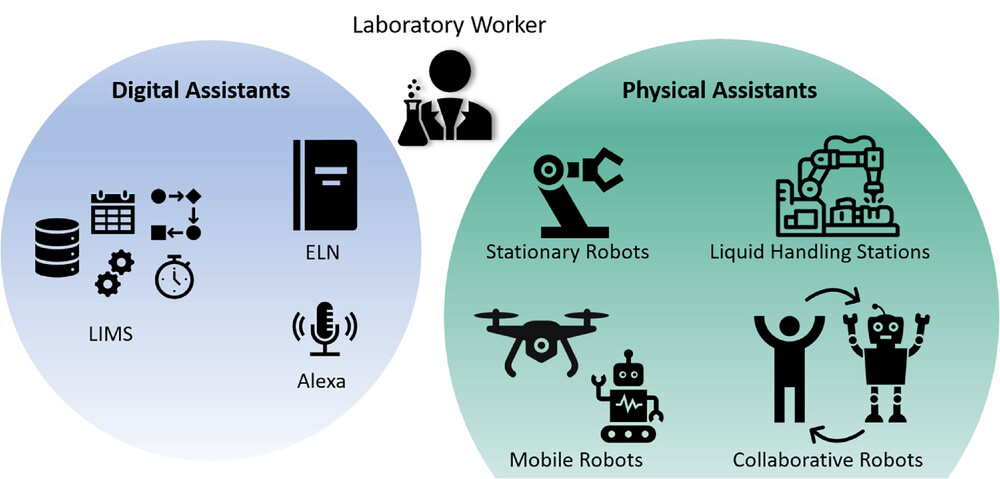
In this 2024 paper published in the journal
Engineering in Life Sciences, Habich and Beutel review the potential technological transformation processes that await the academic bioprocess laboratory, moving from paper-based workflows to electronic ones. Noting multiple challenges facing these academic labs, the authors look at multiple laboratory informatics and automation technologies in the scope of the FAIR (findable, accessible, interoperable, and reusable) data principles, standardized communication protocols, digital twins, and more. After going into detail about these and other related technologies (including electronic laboratory notebooks and laboratory information management systems), the authors present digitalization strategies for academic bioprocess labs. They conclude that "when supported by digital assistants, using connected devices, more intuitive user interfaces, and data gathering according to FAIR data principles, academic research will become less time consuming and more effective and accurate, as well as less prone to (human) error." They add that experiment reproducibility, instrument integration, and other aspect also benefit, though no one-size-fits-all approach exists for these labs.
Posted on April 15, 2024
By LabLynx
Journal articles
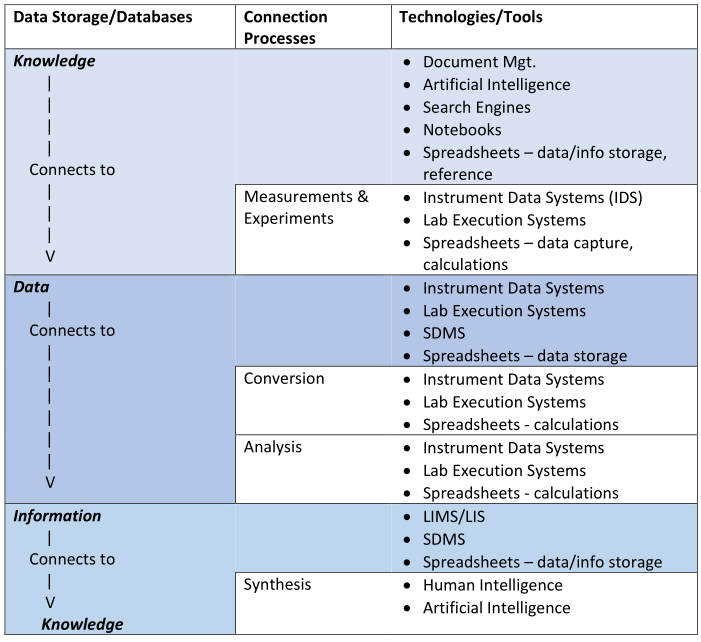
In this 2024 work by laboratory informatics veteran Joe Liscouski, the concept of "organizational memory" within the laboratory realm and what it means for informatics systems like electronic notebooks—including electronic laboratory notebooks (ELNs)—is addressed. Liscouski argues that as artificial intelligence (AI) and large language models (LLMs) become more viable to laboratories, those laboratories stand to benefit from LLM technology in attempts to retain and act upon all sorts of data and information, from raw instrument data and test results to reports and hazardous material records. After addressing what organization memory is and means to the lab, the author discusses the flow of scientific data and information and how it relates to informatics and AI-driven systems. He then discusses the implications as they relate specifically to ELNs and other electronic notebook activities, looking specifically at a layered approach built upon open-source solutions such as Nextcloud "that can feed the [LLM] database as well as support research, development, and other laboratory activities."
Posted on April 10, 2024
By LabLynx
Journal articles
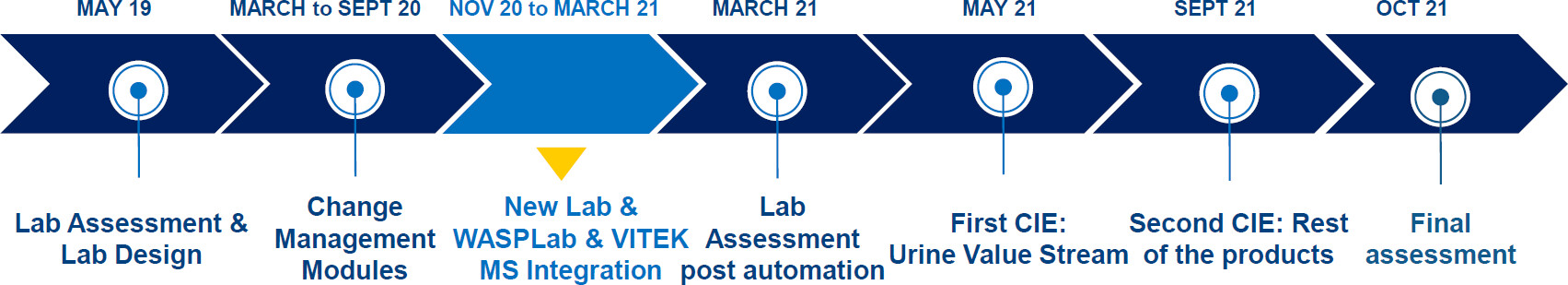
In this 2024 article published in the journal
Microbiology Spectrum, Trigueiro
et al. "explore the relative impact of laboratory automation and continuous improvement events (CIEs) on productivity and [turnaround times (TATs)]" in the clinical microbiology laboratory. Noting numerous advantages to applying the concept of total laboratory automation (TLA), the authors implemented WASPLab and VITEK MS and compared pre-conversion workflow results with post-conversion workflow results. The group also incorporated change management, CIEs, and key performance indicators (KPIs). They concluded that "conversion resulted in substantial improvements in KPIs. While automation alone substantially improved TAT and productivity, the subsequent implementation of lean management further unlocked the potential of laboratory automation through the streamlining of the processes involved." However, they also noted that while effective, some cost is involved with TLA, and labs need "to find ways of maximizing the benefits of the use of [laboratory automation] technology."
Posted on April 1, 2024
By LabLynx
Journal articles
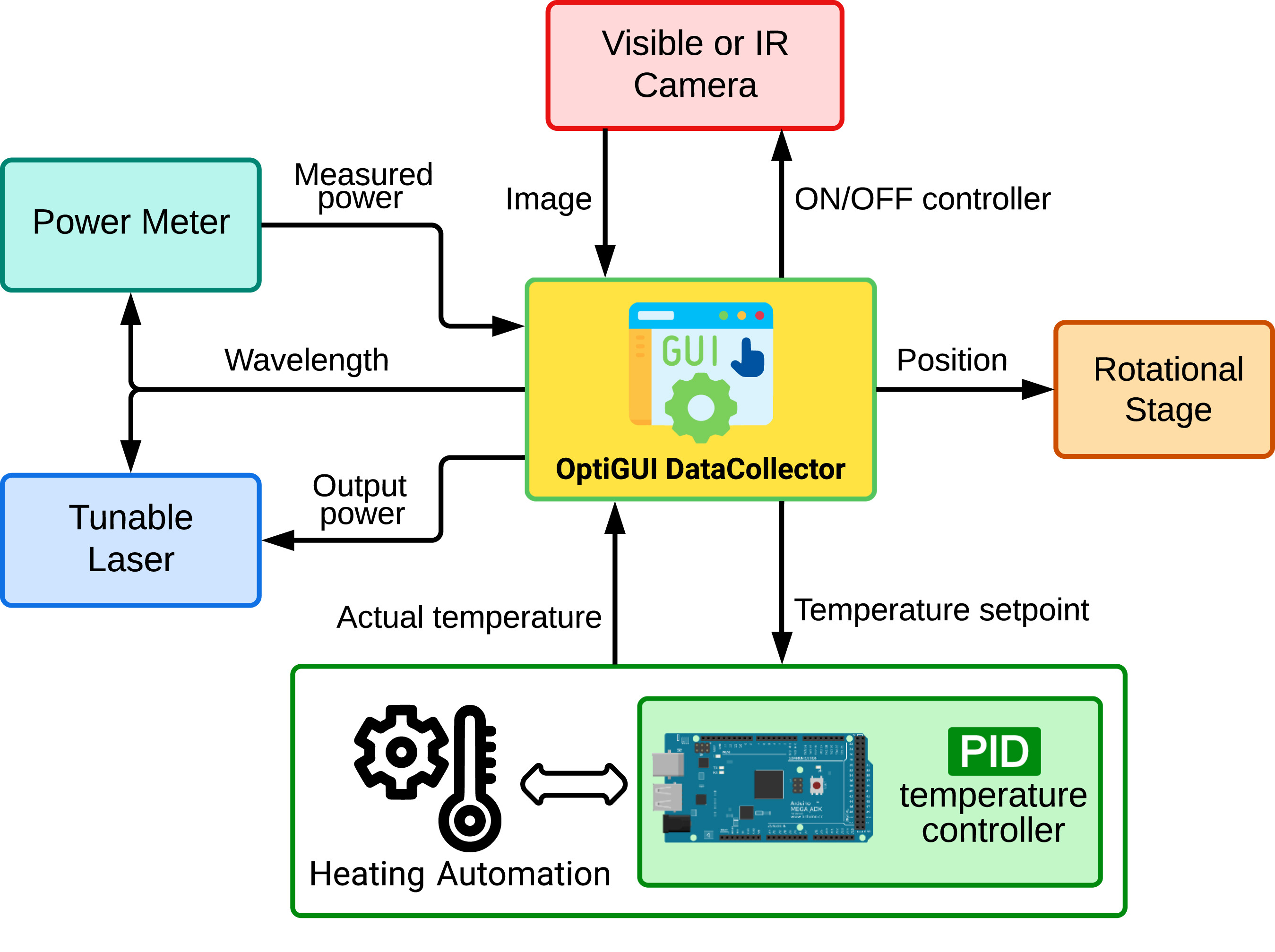
Wanting to integrate instruments and electronic systems in a laboratory is nothing new, and it is in fact increasingly desired. The optics and photonics lab is no exception to this fact. In this 2023 article published in the journal
SoftwareX, Soto-Perdomo
et al. address this desire with their OptiGUI DataCollector, a modular system that integrates optical and photonic devices, simplifies experiment workflows, and automates numerous workflow aspects in the lab, which in turn reduces the number of human-based errors. After a brief introduction to their reasoning for the system, the authors fully describe it and provide illustrative examples of its use. They conclude that the "software's automation capabilities accelerate data acquisition, processing, and analysis, saving researchers time and effort," while also improving reproducibility of research experiments.
Posted on March 26, 2024
By LabLynx
Journal articles

For many years, the journal
PLOS Computational Biology has been publishing a series of "Ten Simple Rules" articles as they relate to data management and sharing in laboratories and research institutes. In this latest installment of the series, Berezin
et al. tap into their collective experience to address the management of laboratory data and information, particularly leveraging systems like a laboratory information management system (LIMS). In these rules, the authors examine aspects such common culture development, inventory management, project management, sample management, labeling, and more. They conclude that apart from "[i]mparting a strong organizational structure for your lab information ... the goal of these rules is also to spur conversation about lab management systems both between and within labs as there is no one-size-fits-all solution for lab management." However, the application of these rules isn't exactly a straightforward process; their success "relies on the successful integration of effective software tools, training programs, lab management policies, and the will to abide by these policies."
Posted on March 18, 2024
By LabLynx
Journal articles
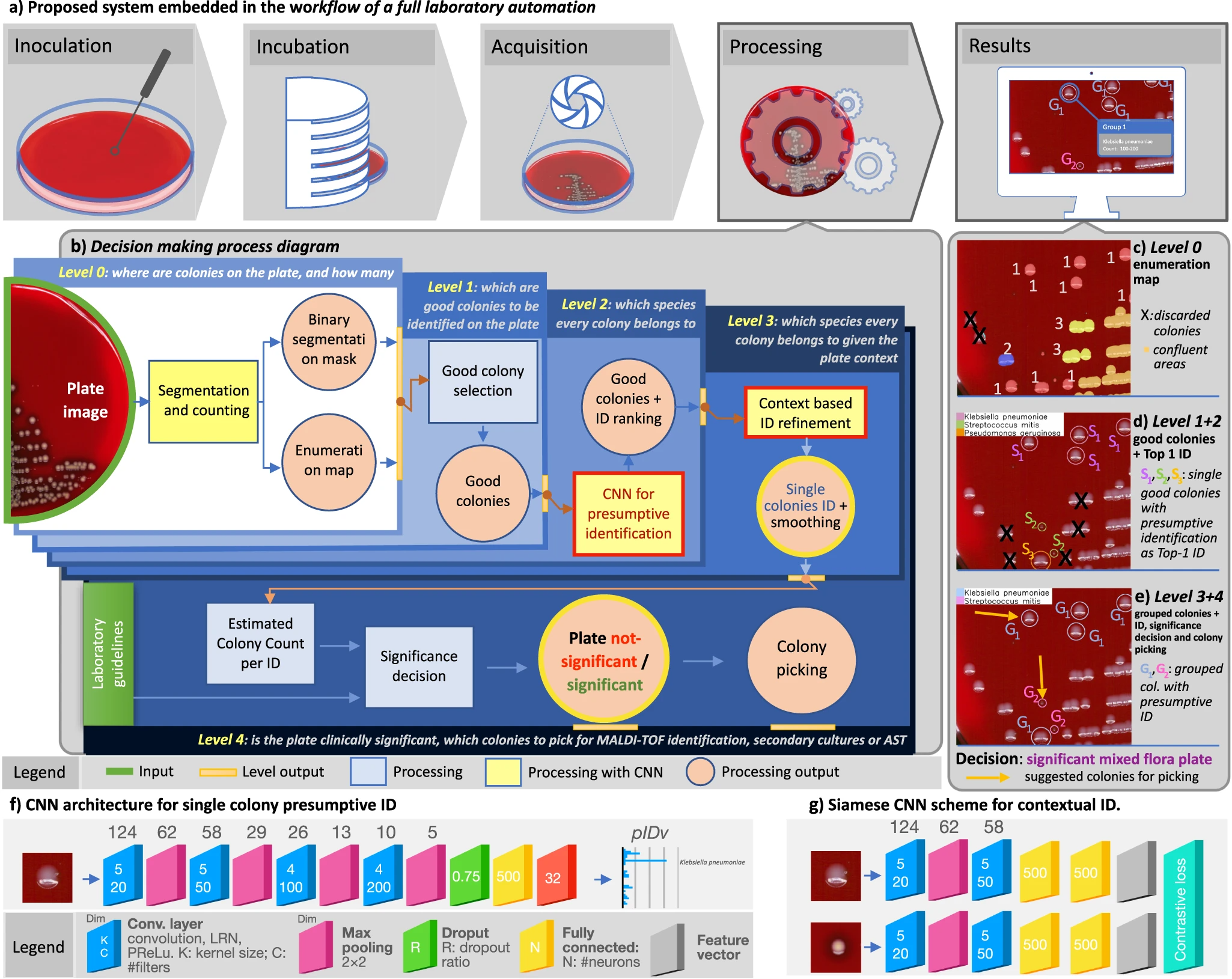
In this 2023 paper published in
Nature Communications, Signoroni
et al. present the results of their efforts towards a system designed for "the global interpretation of diagnostic bacterial culture plates" using deep learning architecture. Noting the many challenges of human-based culture interpretation, the authors present the results of DeepColony," a hierarchical multi-network capable of handling all identification, quantitation, and interpretation stages, from the single colony to the whole plate level." After reviewing the results, the authors conclude that given the "high level of agreement" between DeepColony's results and the interpretation of humans, their system holds significant promise. They add that DeepColony can be viewed as "a unique framework for improving the efficiency and quality of massive routine activities and high-volume decisional procedures in a microbiological laboratory, with great potential to refine and reinforce the critical role of the microbiologist."
Posted on March 11, 2024
By Shawn Douglas
Journal articles
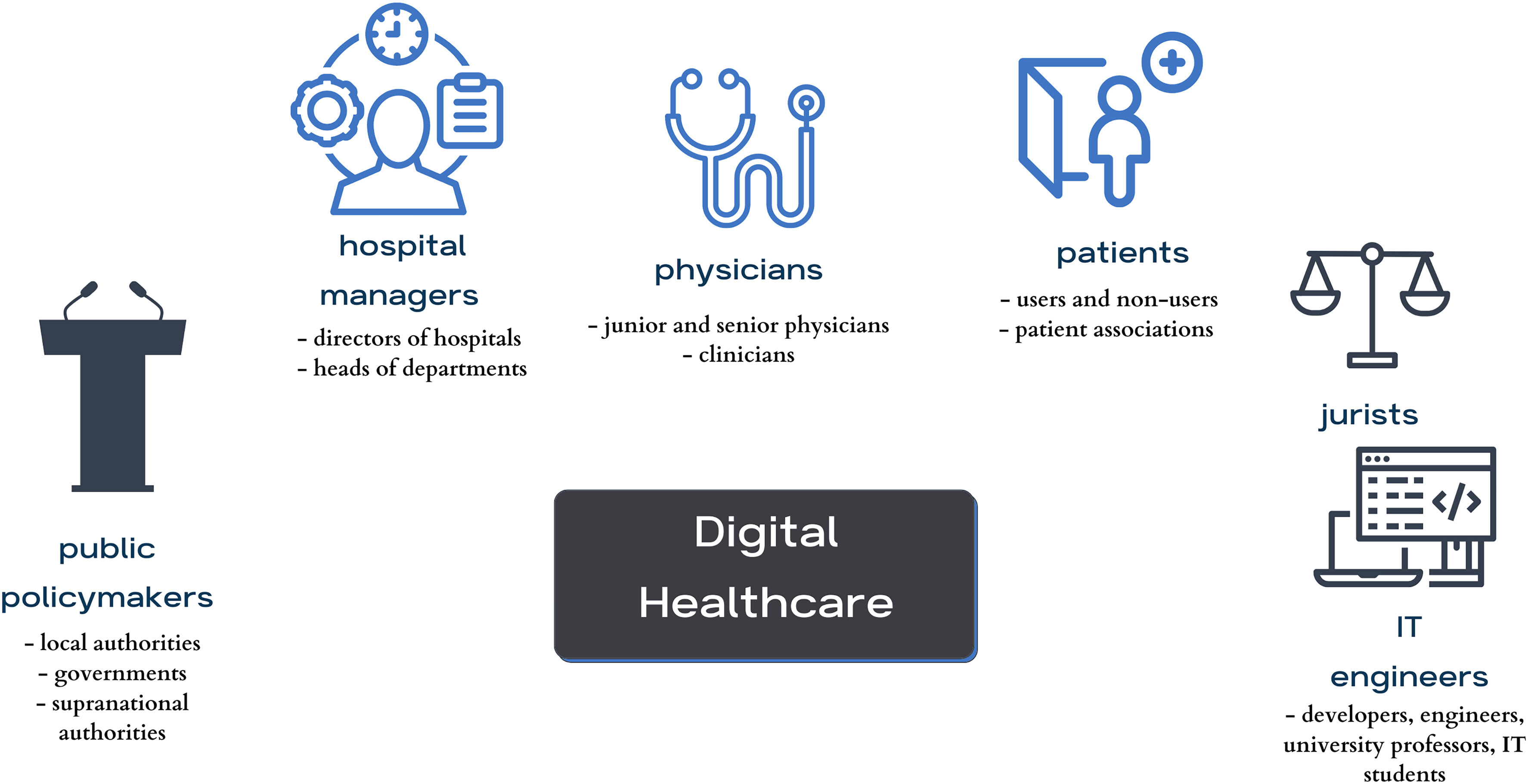
In this 2023 paper published in the journal
Digital Health, Čartolovni
et al. present the results of their qualitative, multi-stakeholder study regarding the "aspirations, expectations, and critical analysis of the potential for artificial intelligence (AI) to transform the patient–physician relationship." Pulling from hours of semi-structured interviews, the authors developed a number of themes from the interviews and discussed how to reach a consensus on their interpretation and use. The authors focused on four main themes and a variety of subthemes and present the results of their work in the framework of these themes and subthemes. From their work, they conclude a definitive "need to use a critical awareness approach to the implementation of AI in healthcare by applying critical thinking and reasoning, rather than simply relying upon the recommendation of the algorithm." Additionally, they highlight how "it is important not to neglect clinical reasoning and consideration of best clinical practices, while avoiding a negative impact on the existing patient–physician relationship by preserving its core values," strongly urging for the preservation of patient-physician trust in the face of increased AI use.
Posted on March 4, 2024
By LabLynx
Journal articles
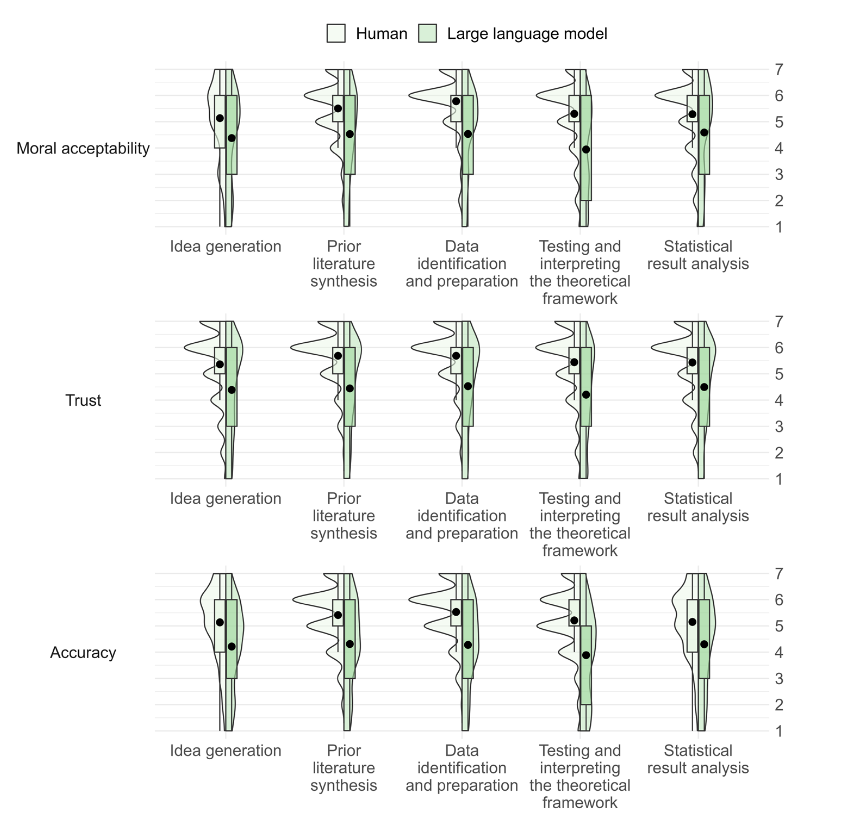
As discussion of artificial intelligence (AI) continues to ramp up in not only academia but also industry and culture, questions abound concerning it's long-term potential and perils. Attitudes about AI and what it means for humanity are diverse, and sometimes controversial. In this 2023 paper published in
Economics and Business Review, Niszczota and Conway contribute to the discussion, particularly in regards to how people view research co-created by generative AI. Recruiting more than 440 individuals, the duo conducted a mixed-design experiment using multiple research processes. All said, their "results suggest that people have clear, strong negative views of scientists delegating any aspect of the research process" to generative AI, denoting it "as immoral, untrustworthy, and scientifically unsound." They conclude that "researchers should employ caution when considering whether to incorporate ChatGPT or other [large language models] into their research."
Posted on February 27, 2024
By LabLynx
Journal articles
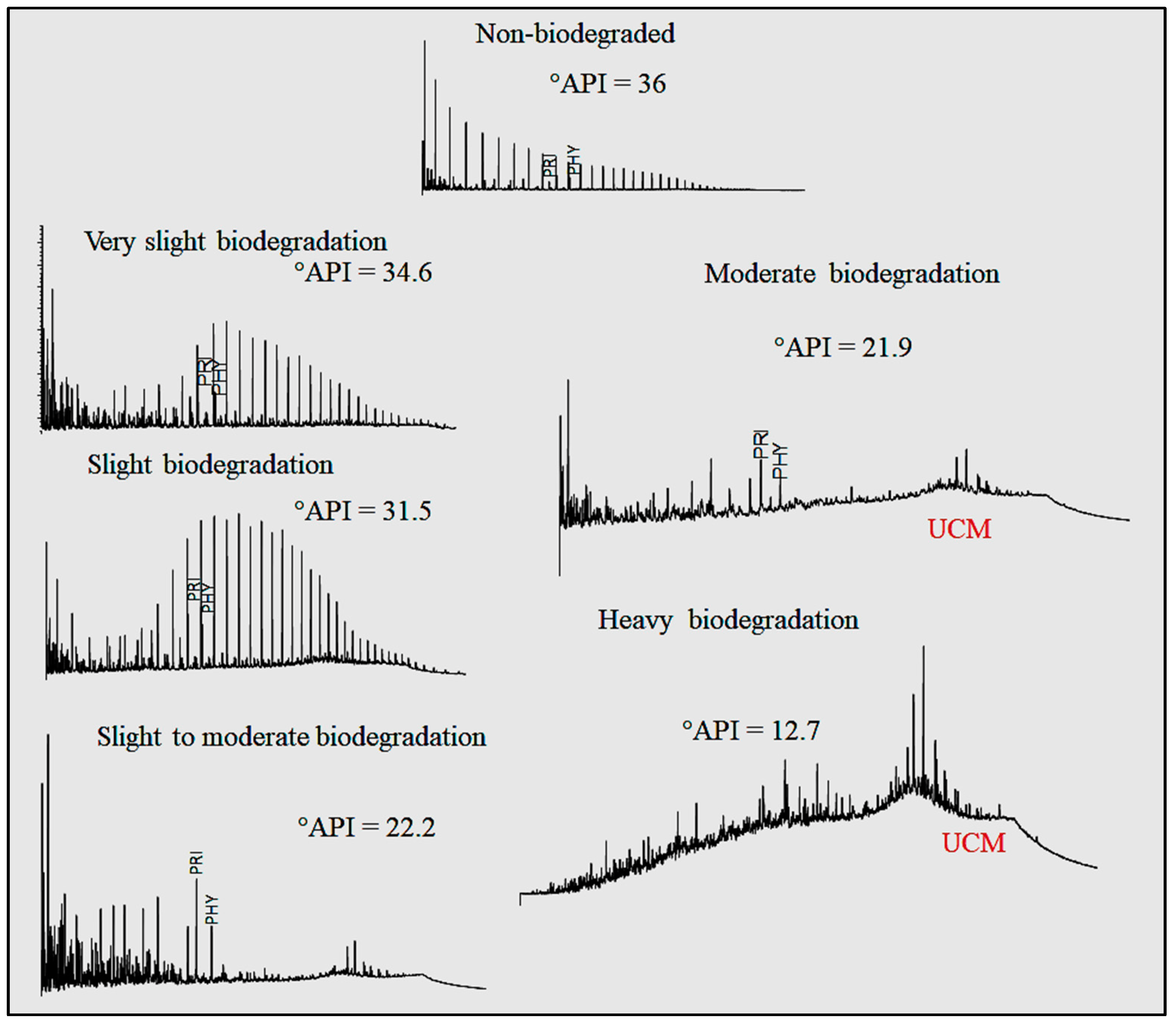
The use of chromatography methods in characterizing and interpreting phenomena related to oil has been around for a long time. However, reading chromatograms—particularly large quantities of them—can be time-comsuming. Bispo-Silva
et al propose a more rapid process using modern deep learning and artificial intelligence (AI) techniques, increasingly being used in large companies. The authors land on using convolutional neural networks (CNN) for this work, particularly in the use of discriminating biodegraded oils from non-biodegraded oils. After presenting background on the topic, as well as their materials and methods, the authors present the results of using CNN and a variety of algorithms to classify chromatograms with both accurate and misleading training materials. The authors conclude that "CNN can open a new horizon for geochemistry when it comes to analysis by" a variety of gas chromatography techniques, as well as "identification of contaminants (as well as environmental pollutants), identification of analysis defects, and, finally, identification and characterization of origin and oil maturation."
Posted on February 20, 2024
By LabLynx
Journal articles
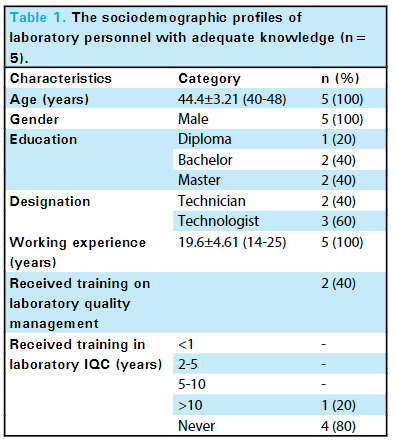
In this brief study by Mishra
et al. in the
Journal of Nepal Medical Association, the level of knowledge of internal quality control (IQC) in the Department of Biochemistry, B.P. Koirala Institute of Health Sciences (BPKIHS), a tertiary care center, is explored. Noting the importance of laboratory quality control and knowledge of IQC to patient outcomes, the authors conducted a descriptive cross-sectional study of its laboratory staff (
n=20), asking questions related to "the understanding of the purpose of IQC, the types of control materials, various control charts, how and when IQC should be performed, and interpretations of the Levey-Jennings Chart using the Westgard rule." The authors concluded from their results were inline with other studies conducted in similar environments (25% had adequate knowledge of IQC), their facility had work to do in improving IQC knowledge and quality management systems more broadly. They add: "Hence, providing training opportunities on laboratory IQC can be reflected as a necessity in our current laboratory set-up. This could add value to the knowledge of IQC on laboratory personnel to ensure that the reports generated within the laboratory are accurate, reliable, and reproducible."
Posted on February 13, 2024
By LabLynx
Journal articles
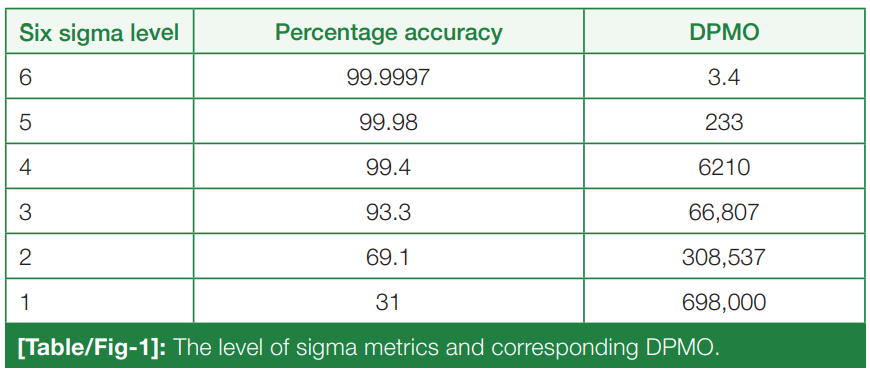
In this 2023 article published in the
National Journal of Laboratory Medicine, Karaattuthazhathu
et al. of KMCT Medical College present the results of a performance assessment of its clinical laboratories' analyte testing parameters using a Six Sigma approach. Examining a six-month period in 2022, the authors looked at 26 parameters in biochemistry and hematology using both internal quality control (IQC) and external quality assurance (EQAS) analyses. After presenting materials and methods used, as well as the results, the authors reviewed their results and concluded that according their sigma metrics analysis, their laboratories are "able to achieve satisfactory results, with world-class performance of many analytes," though recognizing some deficiencies, which were corrected mid-study.
Posted on February 6, 2024
By LabLynx
Journal articles
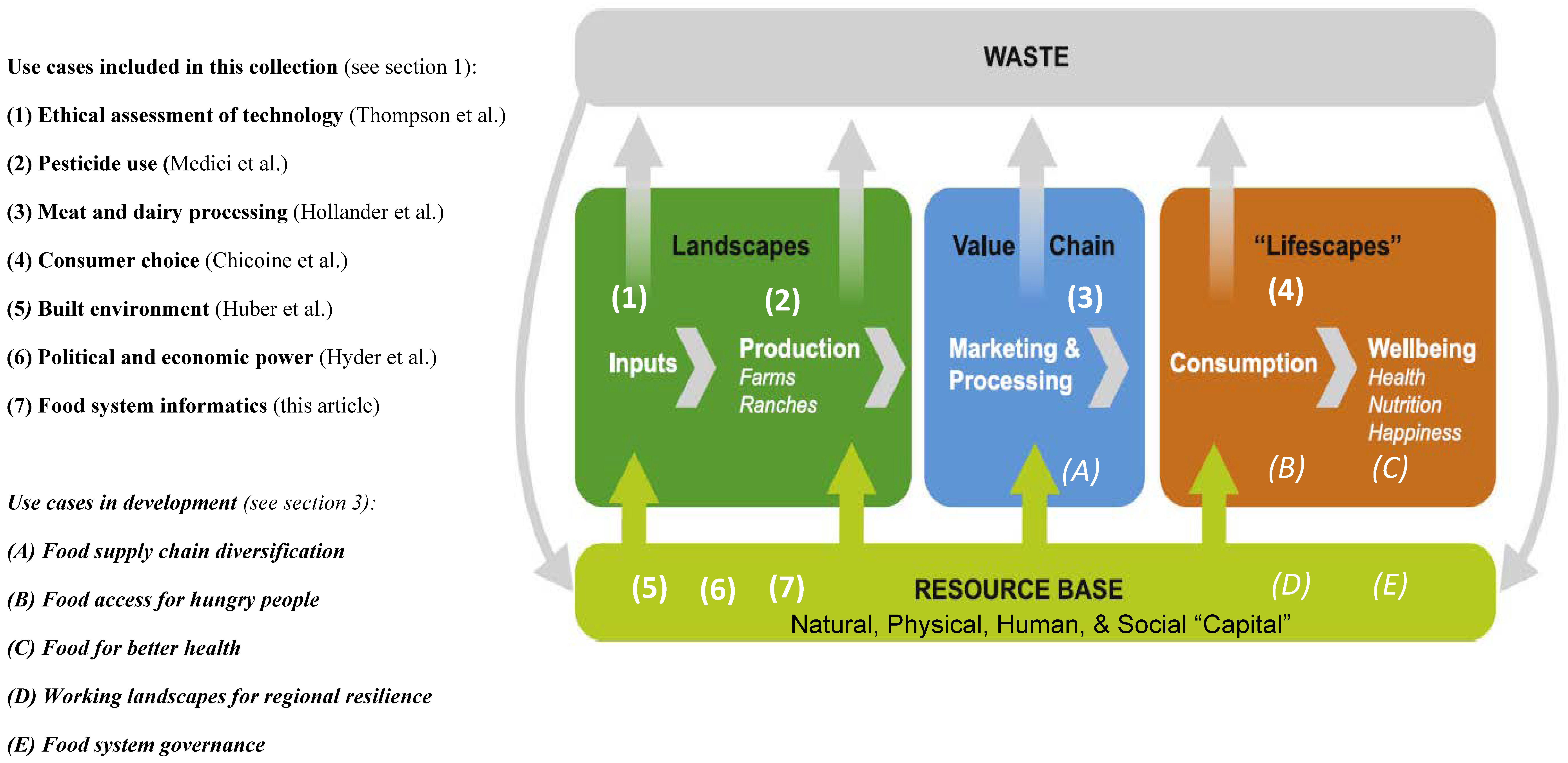
In this 2023 paper published in
Sustainability, Tomich
et al. describe the concept of food systems informatics (FSI) within the context of a collection of this and other paper published as a special collection in the journal. Noting many challenges to improving and transforming food systems, as well as the potential for informatics applications to play an important role, the authors describe five use cases of FSI and discuss the potential outcomes and impacts of developing and implementing FSI platforms in these and other use cases. Finally, the authors draw six major conclusions from their work, as well as several caveats about FSI implementation going forward. They argue that FSI definitely has potential as "a tool to enhance equity, sustainability, and resilience of food systems through collaborative, user-driven interaction, negotiation, experimentation, and innovation within food systems." However, the scope of FSI must be expanded to include " food systems security, privacy, and intellectual property considerations" in order to have the greatest impact.
 In this 2022 paper published in the journal Scientific Data, Deagen et al. present their approach to "web-based, interactive data graphics synchronized to a knowledge graph" in order to make materials science research objects more FAIR (findable, accessible, interoperable, and reusable). After a brief introduction, the authors discuss their methodology and results towards representing charts as metadata, tapping into SPARQL, Vega-Lite, and knowledge graph databases. They also address how their method touches upon multiple aspects of the FAIR data principles. They conclude that "altogether, this pairing of SPARQL and Vega-Lite—demonstrated here in the domain of polymer nanocomposite materials science—offers an extensible approach to FAIR ... scientific data visualization within a knowledge graph framework," adding that "defining charts as metadata in a knowledge graph captures semantic context and visual context while providing interactive, human-interpretable documentation of the contents of a knowledge graph."
In this 2022 paper published in the journal Scientific Data, Deagen et al. present their approach to "web-based, interactive data graphics synchronized to a knowledge graph" in order to make materials science research objects more FAIR (findable, accessible, interoperable, and reusable). After a brief introduction, the authors discuss their methodology and results towards representing charts as metadata, tapping into SPARQL, Vega-Lite, and knowledge graph databases. They also address how their method touches upon multiple aspects of the FAIR data principles. They conclude that "altogether, this pairing of SPARQL and Vega-Lite—demonstrated here in the domain of polymer nanocomposite materials science—offers an extensible approach to FAIR ... scientific data visualization within a knowledge graph framework," adding that "defining charts as metadata in a knowledge graph captures semantic context and visual context while providing interactive, human-interpretable documentation of the contents of a knowledge graph."
 In this 2024 article published in the journal Data, tom Wörden et al. describe their approach to simultaneously using relational database systems with with more semantically driven research data management systems (RDMS) while largely keeping the two in synch. The authors use the open-source RDMS LinkAhead and its associated crawler, as well as YAML for synchronization and data mapping. They describe their approach in detail, noting the importance of "identifiables," which they define as "a set of properties that is taken from the file system layout or the file contents, which allow for uniquely identifying the corresponding object in the RDMS." After a brief discussion of their results and the limitations of their approach, they conclude that the practicality of their approach has been demonstrated, with the potential to transfer "the concept to multiple different use cases in order to make the crawler more robust" across a broader spectrum of scientific research settings.
In this 2024 article published in the journal Data, tom Wörden et al. describe their approach to simultaneously using relational database systems with with more semantically driven research data management systems (RDMS) while largely keeping the two in synch. The authors use the open-source RDMS LinkAhead and its associated crawler, as well as YAML for synchronization and data mapping. They describe their approach in detail, noting the importance of "identifiables," which they define as "a set of properties that is taken from the file system layout or the file contents, which allow for uniquely identifying the corresponding object in the RDMS." After a brief discussion of their results and the limitations of their approach, they conclude that the practicality of their approach has been demonstrated, with the potential to transfer "the concept to multiple different use cases in order to make the crawler more robust" across a broader spectrum of scientific research settings.
 As the ramifications of the FAIR data principles of Wilkinson et al. continue to shape how academic (and industrial) research is performed, it's inevitable that the information management systems used in these research contexts will be scrutinized under those same FAIR data principles. In this case, Plass et al. of FAU Erlangen-Nürnberg describe how they landed upon the open-source electronic laboratory notebook (ELN) and laboratory information management system (LIMS) solution openBIS to address FAIR data management in their multidisciplinary Collaborative Research Centre (CRC). After discussing the current state of research data management (RDM), the authors go into the specifics of their implementation of openBIS, the adjustments they made to the system to address their information management needs, and the experiences they had using the modified system. They ultimately conclude that "this ELN-LIMS could serve as a template solution for other similarly structured collaborative research centers or research groups" given the benefits they saw with their modified version.
As the ramifications of the FAIR data principles of Wilkinson et al. continue to shape how academic (and industrial) research is performed, it's inevitable that the information management systems used in these research contexts will be scrutinized under those same FAIR data principles. In this case, Plass et al. of FAU Erlangen-Nürnberg describe how they landed upon the open-source electronic laboratory notebook (ELN) and laboratory information management system (LIMS) solution openBIS to address FAIR data management in their multidisciplinary Collaborative Research Centre (CRC). After discussing the current state of research data management (RDM), the authors go into the specifics of their implementation of openBIS, the adjustments they made to the system to address their information management needs, and the experiences they had using the modified system. They ultimately conclude that "this ELN-LIMS could serve as a template solution for other similarly structured collaborative research centers or research groups" given the benefits they saw with their modified version.
 In this "in press" journal article published in Practical Laboratory Medicine, Smith et al. of ARUP Laboratories provide the results of a survey conducted just prior to the U.S. Food and Drug Administration's (FDA's) April 29 release of their final rule on laboratory-developed tests (LDTs). Polling a select group of its clinical laboratory customers, the authors sought to discover clinical laboratorians' opinions about the then proposed changes to regulate LDTs as medical devices. The survey was conducted in part due to a set of perceived FDA shortcomings that would make it "difficult for the FDA to conduct an accurate quantitative analysis of public opinion in a key sector directly impacted by its proposed rule." The authors describe the structure of their survey and its associated analytical methods before discussing the results. They then provides a full discussion with a brief conclusion: "The majority of clinical laboratorians surveyed do not support the FDA’s proposed rule on LDTs and report having insufficient resources to comply with the rule if it is enacted," and that the FDA had more work to do "to better understand the negative impacts of the proposed rule."
In this "in press" journal article published in Practical Laboratory Medicine, Smith et al. of ARUP Laboratories provide the results of a survey conducted just prior to the U.S. Food and Drug Administration's (FDA's) April 29 release of their final rule on laboratory-developed tests (LDTs). Polling a select group of its clinical laboratory customers, the authors sought to discover clinical laboratorians' opinions about the then proposed changes to regulate LDTs as medical devices. The survey was conducted in part due to a set of perceived FDA shortcomings that would make it "difficult for the FDA to conduct an accurate quantitative analysis of public opinion in a key sector directly impacted by its proposed rule." The authors describe the structure of their survey and its associated analytical methods before discussing the results. They then provides a full discussion with a brief conclusion: "The majority of clinical laboratorians surveyed do not support the FDA’s proposed rule on LDTs and report having insufficient resources to comply with the rule if it is enacted," and that the FDA had more work to do "to better understand the negative impacts of the proposed rule."
 Histopathology diagnostics has largely been a manual process, with humans examining images of tissues and evaluating them based on knowledge passed down and experience gained. With the age of digital pathology reaching strong realization, new methods are arising, including with the use of artificial intelligence (AI). In this 2024 paper published in Frontiers in Oncology, Doğan and Yılmaz present their AI-based system for image analysis, which taps into both a histogram of oriented gradients (HoG) method and a customized convolutional neural network (CNN) model. After discussing their approach and results, the authors discuss the implications of their results, concluding that their approach could be "considered a good alternative as it performs as well as and better than the more complex models described in the literature." They also say that their results highlight "the indisputable advantages of AI automation and the critical importance of data normalization and interpretation" for any AI-driven effort towards histopathology image analysis, while at the same time noting that the AI's "algorithms must [still] transparently reveal decision-making processes and be understandable by clinical experts."
Histopathology diagnostics has largely been a manual process, with humans examining images of tissues and evaluating them based on knowledge passed down and experience gained. With the age of digital pathology reaching strong realization, new methods are arising, including with the use of artificial intelligence (AI). In this 2024 paper published in Frontiers in Oncology, Doğan and Yılmaz present their AI-based system for image analysis, which taps into both a histogram of oriented gradients (HoG) method and a customized convolutional neural network (CNN) model. After discussing their approach and results, the authors discuss the implications of their results, concluding that their approach could be "considered a good alternative as it performs as well as and better than the more complex models described in the literature." They also say that their results highlight "the indisputable advantages of AI automation and the critical importance of data normalization and interpretation" for any AI-driven effort towards histopathology image analysis, while at the same time noting that the AI's "algorithms must [still] transparently reveal decision-making processes and be understandable by clinical experts."
 In this 2023 article published in the Journal of Applied Clinical Medical Physics, Kapoor et al. of Virginia Commonwealth University present their approach to managing radiation oncology research data in a FAIR (findable, accessible, interoperable, and reusable) way. The authors focus on the advantages of a knowledge graph-based database combined with ontology-based keyword search, synonym-based term matching, and a variety of visualization tools. After presenting background on the topic, Kapoor et al. describe their approach using 1,660 patient clinical and dosimetry records, standardize terminologies, data dictionaries, and Semantic Web technologies. They then discussed the results of their mapping efforts, as well as the discovered benefits of a knowledge graph-based solution over a traditional relational database approach. They conclude that their approach "successfully demonstrates the procedures of gathering data from multiple clinical systems and using ontology-based data integration," with the radiation oncology datasets being more FAIR "using open semantic ontology-based formats," which in turn helps "facilitate interoperability and execution of large scientific studies."
In this 2023 article published in the Journal of Applied Clinical Medical Physics, Kapoor et al. of Virginia Commonwealth University present their approach to managing radiation oncology research data in a FAIR (findable, accessible, interoperable, and reusable) way. The authors focus on the advantages of a knowledge graph-based database combined with ontology-based keyword search, synonym-based term matching, and a variety of visualization tools. After presenting background on the topic, Kapoor et al. describe their approach using 1,660 patient clinical and dosimetry records, standardize terminologies, data dictionaries, and Semantic Web technologies. They then discussed the results of their mapping efforts, as well as the discovered benefits of a knowledge graph-based solution over a traditional relational database approach. They conclude that their approach "successfully demonstrates the procedures of gathering data from multiple clinical systems and using ontology-based data integration," with the radiation oncology datasets being more FAIR "using open semantic ontology-based formats," which in turn helps "facilitate interoperability and execution of large scientific studies."
 Can the thoughtful application and optimization of laboratory informatics solutions lead to do a variety of improvements in a hospital-based emergency medicine laboratory? Zhang et al. sought to answer this question in a a contemporary tertiary second-class general hospital in China. After analyzing their emergency laboratory workflows, identifying problems withing those workflows, and identifying other areas of optimization, the authors applied a laboratory information management system (LIMS) to their operations, upgraded instrumentation, and finessed other aspects of their IT infrastructure. After discussing their results, the authors concluded that "the automation and standardization of most stages in emergency laboratory testing can be realized by IT, which reduces the workload of employees and improves emergency laboratory test quality," further noting that "emergency laboratory test report time can be shortened, emergency laboratory test quality can be enhanced, and employee and patient satisfaction can also be improved."
Can the thoughtful application and optimization of laboratory informatics solutions lead to do a variety of improvements in a hospital-based emergency medicine laboratory? Zhang et al. sought to answer this question in a a contemporary tertiary second-class general hospital in China. After analyzing their emergency laboratory workflows, identifying problems withing those workflows, and identifying other areas of optimization, the authors applied a laboratory information management system (LIMS) to their operations, upgraded instrumentation, and finessed other aspects of their IT infrastructure. After discussing their results, the authors concluded that "the automation and standardization of most stages in emergency laboratory testing can be realized by IT, which reduces the workload of employees and improves emergency laboratory test quality," further noting that "emergency laboratory test report time can be shortened, emergency laboratory test quality can be enhanced, and employee and patient satisfaction can also be improved."
 In this 2024 paper published in the journal Engineering in Life Sciences, Habich and Beutel review the potential technological transformation processes that await the academic bioprocess laboratory, moving from paper-based workflows to electronic ones. Noting multiple challenges facing these academic labs, the authors look at multiple laboratory informatics and automation technologies in the scope of the FAIR (findable, accessible, interoperable, and reusable) data principles, standardized communication protocols, digital twins, and more. After going into detail about these and other related technologies (including electronic laboratory notebooks and laboratory information management systems), the authors present digitalization strategies for academic bioprocess labs. They conclude that "when supported by digital assistants, using connected devices, more intuitive user interfaces, and data gathering according to FAIR data principles, academic research will become less time consuming and more effective and accurate, as well as less prone to (human) error." They add that experiment reproducibility, instrument integration, and other aspect also benefit, though no one-size-fits-all approach exists for these labs.
In this 2024 paper published in the journal Engineering in Life Sciences, Habich and Beutel review the potential technological transformation processes that await the academic bioprocess laboratory, moving from paper-based workflows to electronic ones. Noting multiple challenges facing these academic labs, the authors look at multiple laboratory informatics and automation technologies in the scope of the FAIR (findable, accessible, interoperable, and reusable) data principles, standardized communication protocols, digital twins, and more. After going into detail about these and other related technologies (including electronic laboratory notebooks and laboratory information management systems), the authors present digitalization strategies for academic bioprocess labs. They conclude that "when supported by digital assistants, using connected devices, more intuitive user interfaces, and data gathering according to FAIR data principles, academic research will become less time consuming and more effective and accurate, as well as less prone to (human) error." They add that experiment reproducibility, instrument integration, and other aspect also benefit, though no one-size-fits-all approach exists for these labs.
 In this 2024 work by laboratory informatics veteran Joe Liscouski, the concept of "organizational memory" within the laboratory realm and what it means for informatics systems like electronic notebooks—including electronic laboratory notebooks (ELNs)—is addressed. Liscouski argues that as artificial intelligence (AI) and large language models (LLMs) become more viable to laboratories, those laboratories stand to benefit from LLM technology in attempts to retain and act upon all sorts of data and information, from raw instrument data and test results to reports and hazardous material records. After addressing what organization memory is and means to the lab, the author discusses the flow of scientific data and information and how it relates to informatics and AI-driven systems. He then discusses the implications as they relate specifically to ELNs and other electronic notebook activities, looking specifically at a layered approach built upon open-source solutions such as Nextcloud "that can feed the [LLM] database as well as support research, development, and other laboratory activities."
In this 2024 work by laboratory informatics veteran Joe Liscouski, the concept of "organizational memory" within the laboratory realm and what it means for informatics systems like electronic notebooks—including electronic laboratory notebooks (ELNs)—is addressed. Liscouski argues that as artificial intelligence (AI) and large language models (LLMs) become more viable to laboratories, those laboratories stand to benefit from LLM technology in attempts to retain and act upon all sorts of data and information, from raw instrument data and test results to reports and hazardous material records. After addressing what organization memory is and means to the lab, the author discusses the flow of scientific data and information and how it relates to informatics and AI-driven systems. He then discusses the implications as they relate specifically to ELNs and other electronic notebook activities, looking specifically at a layered approach built upon open-source solutions such as Nextcloud "that can feed the [LLM] database as well as support research, development, and other laboratory activities."
 In this 2024 article published in the journal Microbiology Spectrum, Trigueiro et al. "explore the relative impact of laboratory automation and continuous improvement events (CIEs) on productivity and [turnaround times (TATs)]" in the clinical microbiology laboratory. Noting numerous advantages to applying the concept of total laboratory automation (TLA), the authors implemented WASPLab and VITEK MS and compared pre-conversion workflow results with post-conversion workflow results. The group also incorporated change management, CIEs, and key performance indicators (KPIs). They concluded that "conversion resulted in substantial improvements in KPIs. While automation alone substantially improved TAT and productivity, the subsequent implementation of lean management further unlocked the potential of laboratory automation through the streamlining of the processes involved." However, they also noted that while effective, some cost is involved with TLA, and labs need "to find ways of maximizing the benefits of the use of [laboratory automation] technology."
In this 2024 article published in the journal Microbiology Spectrum, Trigueiro et al. "explore the relative impact of laboratory automation and continuous improvement events (CIEs) on productivity and [turnaround times (TATs)]" in the clinical microbiology laboratory. Noting numerous advantages to applying the concept of total laboratory automation (TLA), the authors implemented WASPLab and VITEK MS and compared pre-conversion workflow results with post-conversion workflow results. The group also incorporated change management, CIEs, and key performance indicators (KPIs). They concluded that "conversion resulted in substantial improvements in KPIs. While automation alone substantially improved TAT and productivity, the subsequent implementation of lean management further unlocked the potential of laboratory automation through the streamlining of the processes involved." However, they also noted that while effective, some cost is involved with TLA, and labs need "to find ways of maximizing the benefits of the use of [laboratory automation] technology."
 Wanting to integrate instruments and electronic systems in a laboratory is nothing new, and it is in fact increasingly desired. The optics and photonics lab is no exception to this fact. In this 2023 article published in the journal SoftwareX, Soto-Perdomo et al. address this desire with their OptiGUI DataCollector, a modular system that integrates optical and photonic devices, simplifies experiment workflows, and automates numerous workflow aspects in the lab, which in turn reduces the number of human-based errors. After a brief introduction to their reasoning for the system, the authors fully describe it and provide illustrative examples of its use. They conclude that the "software's automation capabilities accelerate data acquisition, processing, and analysis, saving researchers time and effort," while also improving reproducibility of research experiments.
Wanting to integrate instruments and electronic systems in a laboratory is nothing new, and it is in fact increasingly desired. The optics and photonics lab is no exception to this fact. In this 2023 article published in the journal SoftwareX, Soto-Perdomo et al. address this desire with their OptiGUI DataCollector, a modular system that integrates optical and photonic devices, simplifies experiment workflows, and automates numerous workflow aspects in the lab, which in turn reduces the number of human-based errors. After a brief introduction to their reasoning for the system, the authors fully describe it and provide illustrative examples of its use. They conclude that the "software's automation capabilities accelerate data acquisition, processing, and analysis, saving researchers time and effort," while also improving reproducibility of research experiments.
 For many years, the journal PLOS Computational Biology has been publishing a series of "Ten Simple Rules" articles as they relate to data management and sharing in laboratories and research institutes. In this latest installment of the series, Berezin et al. tap into their collective experience to address the management of laboratory data and information, particularly leveraging systems like a laboratory information management system (LIMS). In these rules, the authors examine aspects such common culture development, inventory management, project management, sample management, labeling, and more. They conclude that apart from "[i]mparting a strong organizational structure for your lab information ... the goal of these rules is also to spur conversation about lab management systems both between and within labs as there is no one-size-fits-all solution for lab management." However, the application of these rules isn't exactly a straightforward process; their success "relies on the successful integration of effective software tools, training programs, lab management policies, and the will to abide by these policies."
For many years, the journal PLOS Computational Biology has been publishing a series of "Ten Simple Rules" articles as they relate to data management and sharing in laboratories and research institutes. In this latest installment of the series, Berezin et al. tap into their collective experience to address the management of laboratory data and information, particularly leveraging systems like a laboratory information management system (LIMS). In these rules, the authors examine aspects such common culture development, inventory management, project management, sample management, labeling, and more. They conclude that apart from "[i]mparting a strong organizational structure for your lab information ... the goal of these rules is also to spur conversation about lab management systems both between and within labs as there is no one-size-fits-all solution for lab management." However, the application of these rules isn't exactly a straightforward process; their success "relies on the successful integration of effective software tools, training programs, lab management policies, and the will to abide by these policies."
 In this 2023 paper published in Nature Communications, Signoroni et al. present the results of their efforts towards a system designed for "the global interpretation of diagnostic bacterial culture plates" using deep learning architecture. Noting the many challenges of human-based culture interpretation, the authors present the results of DeepColony," a hierarchical multi-network capable of handling all identification, quantitation, and interpretation stages, from the single colony to the whole plate level." After reviewing the results, the authors conclude that given the "high level of agreement" between DeepColony's results and the interpretation of humans, their system holds significant promise. They add that DeepColony can be viewed as "a unique framework for improving the efficiency and quality of massive routine activities and high-volume decisional procedures in a microbiological laboratory, with great potential to refine and reinforce the critical role of the microbiologist."
In this 2023 paper published in Nature Communications, Signoroni et al. present the results of their efforts towards a system designed for "the global interpretation of diagnostic bacterial culture plates" using deep learning architecture. Noting the many challenges of human-based culture interpretation, the authors present the results of DeepColony," a hierarchical multi-network capable of handling all identification, quantitation, and interpretation stages, from the single colony to the whole plate level." After reviewing the results, the authors conclude that given the "high level of agreement" between DeepColony's results and the interpretation of humans, their system holds significant promise. They add that DeepColony can be viewed as "a unique framework for improving the efficiency and quality of massive routine activities and high-volume decisional procedures in a microbiological laboratory, with great potential to refine and reinforce the critical role of the microbiologist."
 In this 2023 paper published in the journal Digital Health, Čartolovni et al. present the results of their qualitative, multi-stakeholder study regarding the "aspirations, expectations, and critical analysis of the potential for artificial intelligence (AI) to transform the patient–physician relationship." Pulling from hours of semi-structured interviews, the authors developed a number of themes from the interviews and discussed how to reach a consensus on their interpretation and use. The authors focused on four main themes and a variety of subthemes and present the results of their work in the framework of these themes and subthemes. From their work, they conclude a definitive "need to use a critical awareness approach to the implementation of AI in healthcare by applying critical thinking and reasoning, rather than simply relying upon the recommendation of the algorithm." Additionally, they highlight how "it is important not to neglect clinical reasoning and consideration of best clinical practices, while avoiding a negative impact on the existing patient–physician relationship by preserving its core values," strongly urging for the preservation of patient-physician trust in the face of increased AI use.
In this 2023 paper published in the journal Digital Health, Čartolovni et al. present the results of their qualitative, multi-stakeholder study regarding the "aspirations, expectations, and critical analysis of the potential for artificial intelligence (AI) to transform the patient–physician relationship." Pulling from hours of semi-structured interviews, the authors developed a number of themes from the interviews and discussed how to reach a consensus on their interpretation and use. The authors focused on four main themes and a variety of subthemes and present the results of their work in the framework of these themes and subthemes. From their work, they conclude a definitive "need to use a critical awareness approach to the implementation of AI in healthcare by applying critical thinking and reasoning, rather than simply relying upon the recommendation of the algorithm." Additionally, they highlight how "it is important not to neglect clinical reasoning and consideration of best clinical practices, while avoiding a negative impact on the existing patient–physician relationship by preserving its core values," strongly urging for the preservation of patient-physician trust in the face of increased AI use.
 As discussion of artificial intelligence (AI) continues to ramp up in not only academia but also industry and culture, questions abound concerning it's long-term potential and perils. Attitudes about AI and what it means for humanity are diverse, and sometimes controversial. In this 2023 paper published in Economics and Business Review, Niszczota and Conway contribute to the discussion, particularly in regards to how people view research co-created by generative AI. Recruiting more than 440 individuals, the duo conducted a mixed-design experiment using multiple research processes. All said, their "results suggest that people have clear, strong negative views of scientists delegating any aspect of the research process" to generative AI, denoting it "as immoral, untrustworthy, and scientifically unsound." They conclude that "researchers should employ caution when considering whether to incorporate ChatGPT or other [large language models] into their research."
As discussion of artificial intelligence (AI) continues to ramp up in not only academia but also industry and culture, questions abound concerning it's long-term potential and perils. Attitudes about AI and what it means for humanity are diverse, and sometimes controversial. In this 2023 paper published in Economics and Business Review, Niszczota and Conway contribute to the discussion, particularly in regards to how people view research co-created by generative AI. Recruiting more than 440 individuals, the duo conducted a mixed-design experiment using multiple research processes. All said, their "results suggest that people have clear, strong negative views of scientists delegating any aspect of the research process" to generative AI, denoting it "as immoral, untrustworthy, and scientifically unsound." They conclude that "researchers should employ caution when considering whether to incorporate ChatGPT or other [large language models] into their research."
 The use of chromatography methods in characterizing and interpreting phenomena related to oil has been around for a long time. However, reading chromatograms—particularly large quantities of them—can be time-comsuming. Bispo-Silva et al propose a more rapid process using modern deep learning and artificial intelligence (AI) techniques, increasingly being used in large companies. The authors land on using convolutional neural networks (CNN) for this work, particularly in the use of discriminating biodegraded oils from non-biodegraded oils. After presenting background on the topic, as well as their materials and methods, the authors present the results of using CNN and a variety of algorithms to classify chromatograms with both accurate and misleading training materials. The authors conclude that "CNN can open a new horizon for geochemistry when it comes to analysis by" a variety of gas chromatography techniques, as well as "identification of contaminants (as well as environmental pollutants), identification of analysis defects, and, finally, identification and characterization of origin and oil maturation."
The use of chromatography methods in characterizing and interpreting phenomena related to oil has been around for a long time. However, reading chromatograms—particularly large quantities of them—can be time-comsuming. Bispo-Silva et al propose a more rapid process using modern deep learning and artificial intelligence (AI) techniques, increasingly being used in large companies. The authors land on using convolutional neural networks (CNN) for this work, particularly in the use of discriminating biodegraded oils from non-biodegraded oils. After presenting background on the topic, as well as their materials and methods, the authors present the results of using CNN and a variety of algorithms to classify chromatograms with both accurate and misleading training materials. The authors conclude that "CNN can open a new horizon for geochemistry when it comes to analysis by" a variety of gas chromatography techniques, as well as "identification of contaminants (as well as environmental pollutants), identification of analysis defects, and, finally, identification and characterization of origin and oil maturation."
 In this brief study by Mishra et al. in the Journal of Nepal Medical Association, the level of knowledge of internal quality control (IQC) in the Department of Biochemistry, B.P. Koirala Institute of Health Sciences (BPKIHS), a tertiary care center, is explored. Noting the importance of laboratory quality control and knowledge of IQC to patient outcomes, the authors conducted a descriptive cross-sectional study of its laboratory staff (n=20), asking questions related to "the understanding of the purpose of IQC, the types of control materials, various control charts, how and when IQC should be performed, and interpretations of the Levey-Jennings Chart using the Westgard rule." The authors concluded from their results were inline with other studies conducted in similar environments (25% had adequate knowledge of IQC), their facility had work to do in improving IQC knowledge and quality management systems more broadly. They add: "Hence, providing training opportunities on laboratory IQC can be reflected as a necessity in our current laboratory set-up. This could add value to the knowledge of IQC on laboratory personnel to ensure that the reports generated within the laboratory are accurate, reliable, and reproducible."
In this brief study by Mishra et al. in the Journal of Nepal Medical Association, the level of knowledge of internal quality control (IQC) in the Department of Biochemistry, B.P. Koirala Institute of Health Sciences (BPKIHS), a tertiary care center, is explored. Noting the importance of laboratory quality control and knowledge of IQC to patient outcomes, the authors conducted a descriptive cross-sectional study of its laboratory staff (n=20), asking questions related to "the understanding of the purpose of IQC, the types of control materials, various control charts, how and when IQC should be performed, and interpretations of the Levey-Jennings Chart using the Westgard rule." The authors concluded from their results were inline with other studies conducted in similar environments (25% had adequate knowledge of IQC), their facility had work to do in improving IQC knowledge and quality management systems more broadly. They add: "Hence, providing training opportunities on laboratory IQC can be reflected as a necessity in our current laboratory set-up. This could add value to the knowledge of IQC on laboratory personnel to ensure that the reports generated within the laboratory are accurate, reliable, and reproducible."
 In this 2023 article published in the National Journal of Laboratory Medicine, Karaattuthazhathu et al. of KMCT Medical College present the results of a performance assessment of its clinical laboratories' analyte testing parameters using a Six Sigma approach. Examining a six-month period in 2022, the authors looked at 26 parameters in biochemistry and hematology using both internal quality control (IQC) and external quality assurance (EQAS) analyses. After presenting materials and methods used, as well as the results, the authors reviewed their results and concluded that according their sigma metrics analysis, their laboratories are "able to achieve satisfactory results, with world-class performance of many analytes," though recognizing some deficiencies, which were corrected mid-study.
In this 2023 article published in the National Journal of Laboratory Medicine, Karaattuthazhathu et al. of KMCT Medical College present the results of a performance assessment of its clinical laboratories' analyte testing parameters using a Six Sigma approach. Examining a six-month period in 2022, the authors looked at 26 parameters in biochemistry and hematology using both internal quality control (IQC) and external quality assurance (EQAS) analyses. After presenting materials and methods used, as well as the results, the authors reviewed their results and concluded that according their sigma metrics analysis, their laboratories are "able to achieve satisfactory results, with world-class performance of many analytes," though recognizing some deficiencies, which were corrected mid-study.
 In this 2023 paper published in Sustainability, Tomich et al. describe the concept of food systems informatics (FSI) within the context of a collection of this and other paper published as a special collection in the journal. Noting many challenges to improving and transforming food systems, as well as the potential for informatics applications to play an important role, the authors describe five use cases of FSI and discuss the potential outcomes and impacts of developing and implementing FSI platforms in these and other use cases. Finally, the authors draw six major conclusions from their work, as well as several caveats about FSI implementation going forward. They argue that FSI definitely has potential as "a tool to enhance equity, sustainability, and resilience of food systems through collaborative, user-driven interaction, negotiation, experimentation, and innovation within food systems." However, the scope of FSI must be expanded to include " food systems security, privacy, and intellectual property considerations" in order to have the greatest impact.
In this 2023 paper published in Sustainability, Tomich et al. describe the concept of food systems informatics (FSI) within the context of a collection of this and other paper published as a special collection in the journal. Noting many challenges to improving and transforming food systems, as well as the potential for informatics applications to play an important role, the authors describe five use cases of FSI and discuss the potential outcomes and impacts of developing and implementing FSI platforms in these and other use cases. Finally, the authors draw six major conclusions from their work, as well as several caveats about FSI implementation going forward. They argue that FSI definitely has potential as "a tool to enhance equity, sustainability, and resilience of food systems through collaborative, user-driven interaction, negotiation, experimentation, and innovation within food systems." However, the scope of FSI must be expanded to include " food systems security, privacy, and intellectual property considerations" in order to have the greatest impact.












