Bioinformatics education in pathology training: Current scope and future direction
| Full article title | Bioinformatics education in pathology training: Current scope and future direction |
|---|---|
| Journal | Cancer Informatics |
| Author(s) | Clay, Michael R.; Fisher, Kevin E. |
| Author affiliation(s) | St. Jude Children’s Research Hospital, Baylor College of Medicine, Texas Children’s Hospital |
| Primary contact | Email: Kevin dot Fisher at bcm dot edu |
| Year published | 2017 |
| Volume and issue | 16 |
| Page(s) | 1–6 |
| DOI | 10.1177/1176935117703389 |
| ISSN | 1176-9351 |
| Distribution license | Creative Commons Attribution-NonCommercial 4.0 International |
| Website | http://insights.sagepub.com/ |
| Download | http://insights.sagepub.com/redirect_file.php (PDF) |
Abstract
Training anatomic and clinical pathology residents in the principles of bioinformatics is a challenging endeavor. Most residents receive little to no formal exposure to bioinformatics during medical education, and most of the pathology training is spent interpreting histopathology slides using light microscopy or focused on laboratory regulation, management, and interpretation of discrete laboratory data. At a minimum, residents should be familiar with data structure, data pipelines, data manipulation, and data regulations within clinical laboratories. Fellowship-level training should incorporate advanced principles unique to each subspecialty. Barriers to bioinformatics education include the clinical apprenticeship training model, ill-defined educational milestones, inadequate faculty expertise, and limited exposure during medical training. Online educational resources, case-based learning, and incorporation into molecular genomics education could serve as effective educational strategies. Overall, pathology bioinformatics training can be incorporated into pathology resident curricula, provided there is motivation to incorporate institutional support, educational resources, and adequate faculty expertise.
Keywords: Bioinformatics, informatics, residency, education, pathology, training
Introduction
Anatomic pathology (AP) and clinical pathology (CP) residents, fellows, and faculty dedicate countless hours in structured training environments equipped with textbooks, scientific literature, and professional expertise to achieve proficiency in the histopathologic diagnosis of disease and/or interpretation of laboratory data. With the emergence of genomics data and clinical data warehouses, laboratory professionals are now tasked with managing, interpreting, and leveraging data of unprecedented complexity. These complex data necessitate that educators rethink the skills and knowledge required for a graduating trainee to practice in the “information age” of diagnostic medicine.
Bioinformatics is defined as the management, acquisition, manipulation, and presentation of complex biological data sets, and clinical informatics is the application of information management in health care to promote safe, efficient, effective, personalized, and responsive care. Given the breadth of these definitions, it is not surprising that defining aspects germane to clinical training is a nontrivial task. In molecular pathology, in particular, these definitions are intricately linked; e.g., computational scripts manipulate raw data such that pathologists can review and interpret for clinical reporting. Here, we highlight the requisite baseline skill set a pathologist should acquire during training to remain facile in bioinformatics and still fulfill the necessary requirements to graduate from accredited pathology training programs. A few key questions, barriers, and proposed solutions to incorporate bioinformatics into general residency education will also be discussed.
Informatics education in pathology residency
The American College of Graduate Medical Education (ACGME) is a private organization that oversees all accredited medical residency training in the United States. Their primary role is to standardize program requirements and provide operational standards for the sponsoring institution, training hospitals, faculty and program directors, program resources, and duty hours.[1] Also, the ACGME determines educational milestones that serve as specialty-specific data to facilitate improvements to curricula and resident performance and demonstrate the effectiveness of graduate medical education in meeting the needs of the public.[2] Sponsoring institutions receive funds from the federal government to cover the costs of training physicians, and ACGME-accredited residents’ and fellows’ salaries are allocated from these monies. Notably, sponsoring institutions must demonstrate compliance with the ACGME’s educational recommendations to maintain accreditation and receipt of the federal funds.
The American Board of Pathology (ABP) partners with the ACGME to ensure that accredited pathology programs provide their trainees with the necessary requisites to become board eligible for the AP, CP, AP/CP, or AP/neuropathology (AP/NP) examinations, and all potential ABP examinees must complete 36 to 48 months of full-time training in an ACGME-accredited pathology program. Each trainee must receive at least 24 months of AP-only or CP-only training or 18 months each of structured AP and CP training. The remaining 12 months is flexible and may include AP, CP, or research rotations (up to six months). Furthermore, AP board-eligible examinees must complete at least 50 autopsies by the time the application for certification is submitted.[3] An example of a typical 48-month AP/CP curriculum is provided in Table 1.
| ||||||||||||||||||||||||||||
For informatics education, the ACGME requires that all AP, CP, and combined AP/CP residents gain exposure to clinical informatics during their pathology training. The informatics educational milestones state that a trainee should be able to explain, discuss, classify, and apply clinical informatics by participating in operational and strategy meetings, troubleshooting with information technology staff, and applying informatics skills to laboratory management and integrative bioinformatics (e.g., aggregate multiple data sources and multiple data analysis services).[4][5] As a result of these recommendations, formal clinical informatics rotations have been widely incorporated into pathology residency training programs over the past several years, and some institutions have implemented clinical informatics fellowship programs to facilitate board certification in this subspecialty.
Barriers to bioinformatics education
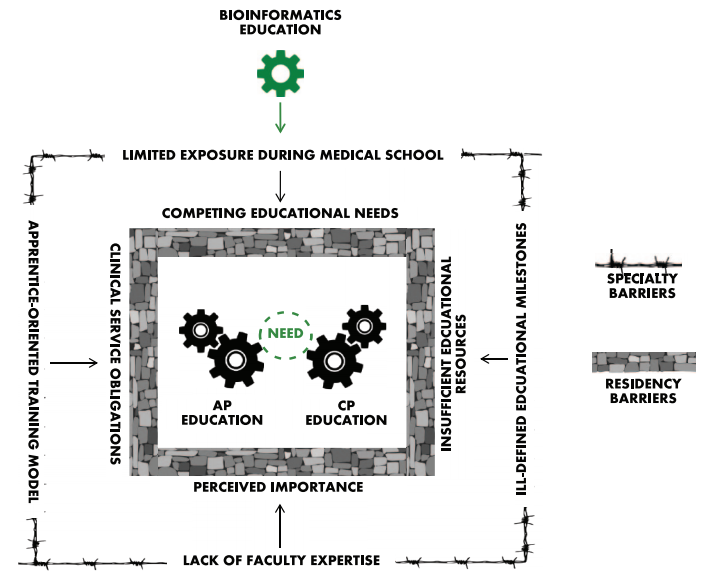
Devising strategies to address bioinformatics education is a complex issue, as there are barriers unique to both the specialty of pathology and residency training in general that must be addressed before bioinformatics can be effectively assimilated into routine pathology education. The key barriers are summarized in Figure 1 and discussed in detail here.
|
The ACGME allows each training program to create and implement its own educational content, and this practice inherently leads to discrepant exposure depending on the demands of other clinical services, faculty expertise, and departmental resources of the individual training programs. Pathology Informatics Essentials for Residents (PIER) is an excellent educational resource that highlights the importance of clinical informatics to the practice of pathology and provides a flexible pathology informatics curriculum and instructional framework that assist training residents in critical pathology informatics knowledge and skills per the ACGME recommended milestones.[6] However, for context, clinical informatics represents only one of the 27 ACGME pathology-specific milestones, and although the PIER topics and educational activities are ideal for instruction in clinical informatics, true bioinformatics topics comprise only a quarter of the 38 clinical informatics master list items. Together, the proposed time allocated to bioinformatics education translates to roughly 0.8% of an entire 48-month AP/CP residency training, or at most 10 days.
The combination of ill-defined educational milestones and limited allocated training time has attributed to underdeveloped and non-standardized bioinformatics educational materials for pathology residents. Ideally, the abovementioned resources could be supplemented with patient-centered, case-based learning modules that would allow trainees the opportunity to work through relevant clinical scenarios. Case-based materials are widely used for training residents in molecular genomic pathology[7] and clinical informatics.[8] However, we are currently unaware of standardized case-based learning materials that are specific to bioinformatics.
Fundamentally, residents “learn their trade” in an apprenticeship model under the tutelage of faculty with expertise in the practice and science of medicine, and as clinical service providers, residents “learn by doing.” For example, in pathology, AP residents gross surgical specimens and extensively study hematoxylin and eosin slides under the light microscope to formulate independent histopathologic diagnoses that are discussed directly with the supervising faculty. Likewise, CP residents work with faculty in secondary roles to interpret serum protein electrophoresis gels, manage transfusion reactions, or a host of other patient-directed finite tasks. Ultimately, this training model dominates almost all medical residency education because it seamlessly integrates trainees into patient care roles that align with the educational mission of the ACGME and the clinical and financial missions of the sponsoring institutions. As a result, residents staff and manage clinical services preferentially over other educational endeavors.
However, bioinformatics education is ill-suited for the clinical service apprenticeship model. Bioinformatics is not a clinical service that residents can receive bioinformatics cases, work up those cases, generate clinical reports, discuss and finalize the reports with the attending (e.g., “sign-out”), bill the patient for clinical services, and repeat the process the next day. Bioinformatics is a data-based discipline where the scope of work is anchored in application projects that may take weeks, months, or even years to materialize in any meaningful manner. Furthermore, the apprentice model requires experienced faculty which may or may not be readily accessible for all pathology trainees. Clinical faculty with limited bioinformatics knowledge, experience, or training may not recognize the importance of bioinformatics education and are therefore more likely to prioritize educational efforts toward clinical services. It is also impractical to assume that one or two informatics faculty members can train all residents in bioinformatics principles, particularly given the aforementioned limited time available (10 days).
There are other systematic factors that impede the incorporation of bioinformatics education into standard pathology curricula. In recent years, medical schools have transitioned from traditional discipline-based didactic curricula to problem-based learning curricula[9][10], most of which offer limited structured informatics education.[11] Furthermore, as an unintended consequence, the principles of histopathology and laboratory medicine were marginalized to electives or abandoned entirely.[12][13] Thus, medical school graduates not only lack exposure to basic bioinformatics principles but also lack exposure to basic principles in pathology which are necessary to function as a competent pathology resident. Therefore, basic histopathology and laboratory medicine principles immediately arise as competing educational needs even at the earliest stages of training.
Although the barriers to bioinformatics education are apparent, clear solutions to address the barriers are not. Given the breadth of diverse information that each resident must obtain during his or her limited time, the limited fund of base knowledge, and the practical consideration that residents are granted 10 days to gain exposure to bioinformatics, important questions remain: What core bioinformatics principles does a general pathologist need to know? What components are best suited for specialized fellowship training? What are successful strategies to implementation? By addressing these questions, a working curriculum for bioinformatics education in pathology residency can be established.
Bioinformatics: What every pathologist needs to know
Each pathologist’s individual practice setting will dictate the breadth and depth of bioinformatics acumen required, but there are core bioinformatics principles that pervade all practices of pathology. Foremost of these is data structure, as a working knowledge allows for effective communication within the health care system. Oftentimes, pathologists are tasked with gathering and assimilating laboratory data to answer clinical questions during multidisciplinary discussions. An understanding of file formats, data organization, and data storage can greatly improve the efficiency of this task, particularly when searching for specific elements that are buried within complex data sets. As a corollary, understanding data structures facilitates collaboration with bioinformaticians such that the data organization is optimized for routine query by the end user.
Understanding the components of bioinformatics pipelines is also important for any pathologist. Most pathologists, particularly those in private practice, are medical directors or involved in professional oversight of at least one clinical laboratory. All clinical data that post to a patient’s permanent medical record are the medical and legal responsibility of the medical director, and clinical laboratories invariably use bioinformatics to manage, acquire, manipulate, and/or present the data. Therefore, by extension, any bioinformatics used in this process are also under the directors’ purview. The pathologist should be familiar with each component of the pipeline so that he or she can understand the flow of information, optimize and troubleshoot as needed, and facilitate technical support for expedited resolution of any issues.
Next-generation sequencing (NGS) workflows in the clinical molecular pathology lab are ideally suited to introduce residents to data structure, file formats, and bioinformatics pipelines. Formal recommended strategies for educating residents in molecular pathology are discussed in detail in several recent publications.[14][15] However, the details of bioinformatics education are varied as some authors have elected to defer to other working groups tasked with specifically addressing informatics education in pathology residencies.[16] Knowledge of how data are generated, processed, and stored provides the learner with insight into the complexities, errors, and pitfalls inherent to the molecular pathology testing cycle and the analytic factors that affect turnaround time. Residents can also gain exposure to testing parameters such as depth of coverage, strand bias, and quality scores, all of which are generated by computational means and are relevant in determining the utility of a particular data set.
Perhaps, more importantly, the identification, classification, and interpretation of genomic alterations function as practical bioinformatics educational material. When classifying an oncologic genomic variant, the pathologist is typically the “end user” who interacts solely with filtered data (e.g., a variant call file) to ascertain clinical therapeutic, diagnostic, or prognostic significance of the variants.[17] Residents should familiarize themselves with how data are filtered prior to receipt by the end user. Also, residents should understand how queries to internal or public databases (e.g., COSMIC [Catalogue of Somatic Mutations in Cancer]) are performed, what databases are available to classify variants as pathogenic, and how databases are populated and curated. Finally, residents should understand how bioinformatics assists with medical context. This includes, but is not limited to, how data such as patient’s history, medical condition, clinical indication, published literature, and many other factors are presented during the variant review process. As a secondary benefit, leveraging bioinformatics for improved patient data review makes excellent resident-appropriate quality assurance projects.
Genomic data may be uniquely identifiable to an individual and are therefore considered personal health information (PHI) according to the Health Insurance Portability and Accountability Act (HIPAA). It is the responsibility of the medical director to physically and electronically safeguard PHI regardless if the data are stored on-site or in HIPAA-compliant secured servers. HIPAA’s three-part security rule provides standards for technical safeguards (data encryption, controlled access, and auditability), physical safeguards (chain of custody, workstation restrictions, mobile device management, facility security, hardware inventory), and administration safeguards (conducting risk assessments, implementation of a risk management policy, employee training, contingency planning, and restricting third-party access). Residents therefore must understand how genomic data are physically, technically, and administratively protected, particularly when third-party software are used during data analysis. This list is not exhaustive, and additional information regarding security rules and implementation can be reviewed at the following website: https://www.hhs.gov/hipaa/for-professionals/security/guidance/index.html.
Advanced training opportunities in bioinformatics
Virtually, all graduating residents pursue advanced training in a pathology subspecialty (e.g., hematopathology, NP, and cytopathology), but few if any informatics topics are listed as ACGME-designated milestone learning objectives for subspecialty training programs.[2] Even in subspecialties where formal bioinformatics educational milestones seem apropos (e.g., molecular genetic pathology and clinical chemistry) the milestones are ill-defined or absent. The absence of milestones often leads to inconsistent self-directed learning and few opportunities for objective assessment. Considering that in 2015 only 21 of 653 (3.2%) subspecialty board examinations were administered in clinical informatics[18], it is clear that most pathologists do not receive structured advanced training in informatics.
Despite the absence of ACGME-defined milestones, fellowship training affords the trainee opportunities for unique, subspecialty-focused bioinformatics exposure. It is these authors’ opinion that until formal guidelines can be determined, each fellowship program/institution must tailor the bioinformatics learning objectives to the institutional mission, bioinformatics needs, and/or the expertise of the faculty and fellows. For example, at Baylor College of Medicine during our annual program evaluation for the molecular genetic pathology fellowship, we identified molecular bioinformatics exposure as an educational improvement area. Each fellow is now required to oversee the validation of a DNA sequencing, RNA sequencing, or copy-number assessment pipeline for all current and future genomic assays. These objectives were pertinent to the clinical mission of the section and combined well with the available faculty expertise. Ongoing discussions at the level of the ACGME and fellowship directors must continue to harmonize informatics education across fellowship programs.
It is worth mentioning that although the leveraging of bioinformatic resources currently centers around the use of NGS in somatic and germline testing, a number of other exciting modalities are on the horizon such as proteomics, metabolomics, and epigenomics[19] that will ultimately integrate into clinical testing and management algorithms. Proteomics, in particular, which is considered the bridge between genomics and biology, has already entered the clinical laboratory via the mass spectrophotometric analysis of microbial agents.[20] It is imperative that subspecialty training programs also strategize how to incorporate these modalities and educate trainees given the inevitable arrival of these complementary data sets.
Bioinformatics education: Strategies to implementation
Clearly, apprentice-based bioinformatics education is suboptimal, and one should not expect proportional educational yield using this method. The absence of extensive in-house expertise and the paucity of finite, readily completable tasks require that educators prioritize bioinformatics concepts and their applications. Online content designed by leading bioinformaticians covers a range of topics from basic computational principles up to BaseSpace and cloud computing coding (Coursera offered by the University of California San Diego[21]) as well as sequencing and quality control, NGS and RNA-seq, R scripting, and basic principles of UNIX operating systems (available from the Boyce Thompson Institute[22]). The Galaxy project[23], a joint venture from Penn State and Johns Hopkins University, is an open-source, web-based platform for data-intensive biomedical research that offers tutorials for users interested in learning about and developing web-based data analysis tools. These are just a few examples of the myriad of available online resources.
We also highlight the pressing need for the development of educational materials. The lack of effective bioinformatics educational material is clearly a barrier to effective implementation (Figure 1), but case studies specifically relating bioinformatics to patient care could easily serve as effective teaching modalities. As the field continues to grow, a collaborative effort with institutions sharing de-identified training materials should be a tangible educational goal. By leveraging these resources, bioinformatics principles can be defined and discussed in the context of ongoing institutional projects. This approach may not necessarily provide comprehensive educational content but can provide a framework for how bioinformatics is used in clinical practice.
Conclusions
Educating residents in the principles of bioinformatics is a daunting challenge alone, and it is made more difficult by the lack of formal guidance from regulatory agencies and the competing educational needs of each pathology residency. All residents should be familiar with data structure, data pipelines, how clinical laboratories manipulate data, and the regulations governing data. Also, a working knowledge of file formats is essential to genomics given the intricate association with the complex genomic data and its application to the diagnosis, prognosis, and predictive clinical management. Finally, bioinformatics principles overlap considerably with clinical informatics and other pathology disciplines that manage, interpret, and use laboratory data routinely. This provides ample opportunities for correlative, clinically directed education on these rotations.
There are numerous online resources available for those interested in designing advanced curricula beyond the basic tenants discussed here. Ideally, bioinformatics should be taught in a concept-based and case-based format, with an emphasis on the applicability to the clinical practice of pathology. Ultimately, however, in order for bioinformatics education to firmly integrate into the fabric of resident education, its importance and broad application to the practice of pathology must be recognized and given a prominent seat at the education table. The stereotype of the traditional diagnostician must be challenged, and bioinformatics will be at the forefront of the renaissance.
Acknowledgements
Author contributions
MRC and KEF conceived and wrote the manuscript. All authors reviewed and approved the final manuscript.
Funding
The author(s) received no financial support for the research, authorship, and/or publication of this article.
Disclosures and ethics
As a requirement of publication, author(s) have provided to the publisher signed confirmation of compliance with legal and ethical obligations including but not limited to the following: authorship and contributorship, conflicts of interest, privacy and confidentiality, and (where applicable) protection of human and animal research subjects. The authors have read and confirmed their agreement with the ICMJE authorship and conflict of interest criteria. The authors have also confirmed that this article is unique and not under consideration or published in any other publication, and that they have permission from rights holders to reproduce any copyrighted material. Any disclosures are made in this section. The external blind peer reviewers report no conflicts of interest.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- ↑ "ACGME Program Requirements for Graduate Medical Education in Anatomic Pathology and Clinical Pathology" (PDF). Accreditation Council for Graduate Medical Education. July 2017. https://www.acgme.org/Portals/0/PFAssets/ProgramRequirements/300_pathology_2017-07-01.pdf. Retrieved 09 August 2017.
- ↑ 2.0 2.1 "Pathology - Milestones". Accreditation Council for Graduate Medical Education. https://www.acgme.org/Specialties/Milestones/pfcatid/18/Pathology. Retrieved 31 October 2016.
- ↑ "Combined Anatomic Pathology and Clinical Pathology (AP/CP)". The American Board of Pathology. http://www.abpath.org/index.php/to-become-certified/requirements-for-certification?layout=edit&id=156. Retrieved 13 January 2017.
- ↑ Naritoku, W.Y.; Alexander, C.B. (2014). "Pathology milestones". Journal of Graduate Medical Education 6 (1 Suppl 1): 180–181. doi:10.4300/JGME-06-01s1-10. PMC PMC3966611. PMID 24701281. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3966611.
- ↑ Naritoku, W.Y.; Alexander, C.B.; Bennett, B.D. et al. (2014). "The pathology milestones and the next accreditation system". Archives of Pathology & Laboratory Medicine 138 (3): 307–15. doi:10.5858/arpa.2013-0260-SA. PMID 24576024.
- ↑ Henricks, W.H.; Karcher, D.S.; Harrison, J.H. et al. (2016). "Pathology Informatics Essentials for Residents: A flexible informatics curriculum linked to Accreditation Council for Graduate Medical Education milestones". Journal of Pathology Informatics 7: 27. doi:10.4103/2153-3539.185673. PMC PMC4977974. PMID 27563486. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4977974.
- ↑ Schrijver, I., ed. (2011). Diagnostic Molecular Pathology in Practice. Springer-Verlag Berlin Heidelberg. pp. 339. doi:10.1007/978-3-642-19677-5. ISBN 9783642196768.
- ↑ Hassell, L.A.; Blick, K.E. (2016). "Training in Informatics: Teaching Informatics in Surgical Pathology". Clinics in Laboratory Medicine 36 (1): 183-97. doi:10.1016/j.cll.2015.09.014. PMID 26851672.
- ↑ Polyzois, I.; Claffey, N.; Mattheos, N. (2010). "Problem-based learning in academic health education: A systematic literature review". European Journal of Dental Education 14 (1): 55-64. doi:10.1111/j.1600-0579.2009.00593.x. PMID 20070800.
- ↑ Burgess, A.W.; McGregor, D.M.; Mellis, C.M. (2014). "Applying established guidelines to team-based learning programs in medical schools: A systematic review". Academic Medicine 89 (4): 678–88. doi:10.1097/ACM.0000000000000162. PMC PMC4885587. PMID 24556770. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4885587.
- ↑ Banerjee, R.; George, P.; Priebe, C.; Alper, E. (2015). "Medical student awareness of and interest in clinical informatics". JAMIA 22 (e1): e42–7. doi:10.1093/jamia/ocu046. PMID 25726567.
- ↑ Smith, B.R.; Kamoun, M.; Hickner, J. (2016). "Laboratory Medicine Education at U.S. Medical Schools: A 2014 Status Report". Academic Medicine 91 (1): 107–12. doi:10.1097/ACM.0000000000000817. PMC PMC5480607. PMID 26200574. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5480607.
- ↑ Laposata, M. (2016). "Insufficient Teaching of Laboratory Medicine in US Medical Schools". Academic Pathology 3: 1–2. doi:10.1177/2374289516634108. PMC PMC5497899. PMID 28725761. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5497899.
- ↑ Aisner, D.L.; Berry, A.; Dawson, D.B. et al. (2016). "A Suggested Molecular Pathology Curriculum for Residents: A Report of the Association for Molecular Pathology". Journal of Molecular Diagnostics 18 (2): 153-62. doi:10.1016/j.jmoldx.2015.10.006. PMID 26857063.
- ↑ Schrijvner, I.; Natkunam, Y.; Galli, S.; Boyd, S.D. (2013). "Integration of genomic medicine into pathology residency training: The Stanford Open Curriculum". Journal of Molecular Diagnostics 15 (2): 141-8. doi:10.1016/j.jmoldx.2012.11.003. PMID 23313248.
- ↑ Laudadio, J.; McNeal, J.L.; Boyd, S.D. et al. (2015). "Design of a Genomics Curriculum: Competencies for Practicing Pathologists". Archives of Pathology & Laboratory Medicine 139 (7): 894-900. doi:10.5858/arpa.2014-0253-CP. PMID 26125429.
- ↑ Li, M.M.; Datto, M.; Duncavage, E.J. et al. (2017). "Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists". Journal of Molecular Diagnostics 19 (1): 4-23. doi:10.1016/j.jmoldx.2016.10.002. PMID 27993330.
- ↑ American Board of Pathology (2015). "Examination Performance" (PDF). The ABP Examiner 37 (2): 9–10. http://www.abpath.org/images/newsletters/2015-2ABPExaminer9_19_16.pdf.
- ↑ Caie, P.D.; Harrison, D.J. (2016). "Next-Generation Pathology". Methods of Molecular Biology 1386: 61–72. doi:10.1007/978-1-4939-3283-2_4. PMID 26677179.
- ↑ van Belkum, A.; Welker, M.; Erhard, M.; Chatellier, S. (2012). "Biomedical mass spectrometry in today's and tomorrow's clinical microbiology laboratories". Journal of Clinical Microbiology 50 (5): 1513-7. doi:10.1128/JCM.00420-12. PMC PMC3347093. PMID 22357505. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3347093.
- ↑ "Coursera - UC San Diego". Coursera, Inc. https://www.coursera.org/ucsd/. Retrieved 31 October 2016.
- ↑ "BTI Plant Bioinformatics Course". Boyce Thompson Institute. https://btiplantbioinfocourse.wordpress.com/. Retrieved 31 October 2016.
- ↑ "Galaxy". The Galaxy Project. https://usegalaxy.org/. Retrieved 31 October 2016.
Notes
This presentation is faithful to the original, with only a few minor changes to presentation, grammar, and spelling. In some cases important information was missing from the references, and that information was added. In one case (the first citation), the original citation URL was dead, and an updated URL was substituted.