Posted on May 16, 2016
By John Jones
Journal articles
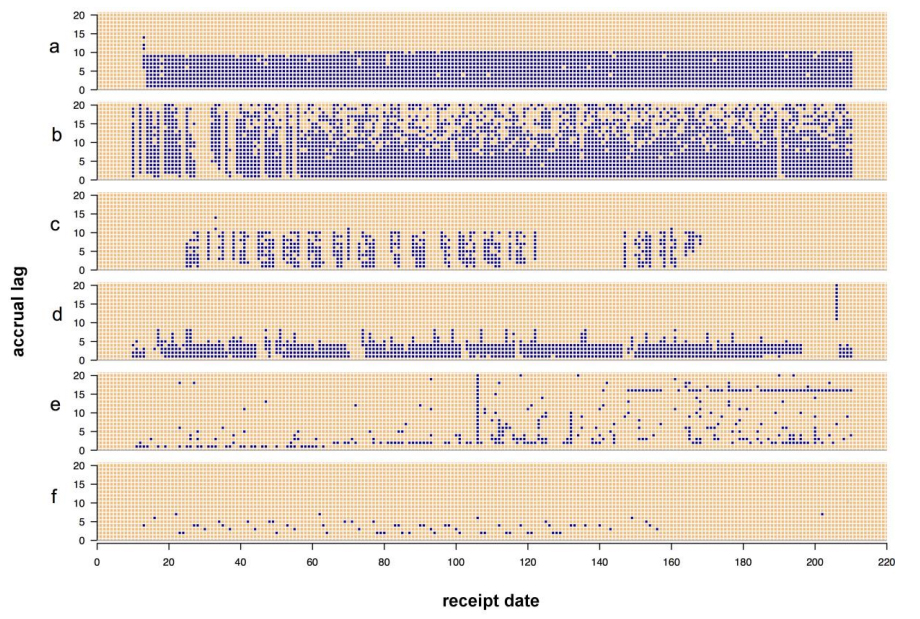
The state of data management across the sciences is getting increasingly complex as data stores build up, and the world of public health is no less affected. Making sense of data is one portion of management, but quality analysis is also an important but slightly understated aspect as well. This 2015 paper by Eaton
et al. explains a series of "data quality tools developed to gain insight into the data quality problems associated with these data." The group concludes "our key insight was the need to assess temporal patterns in the data in terms of accrual lag."
Posted on May 9, 2016
By John Jones
Journal articles
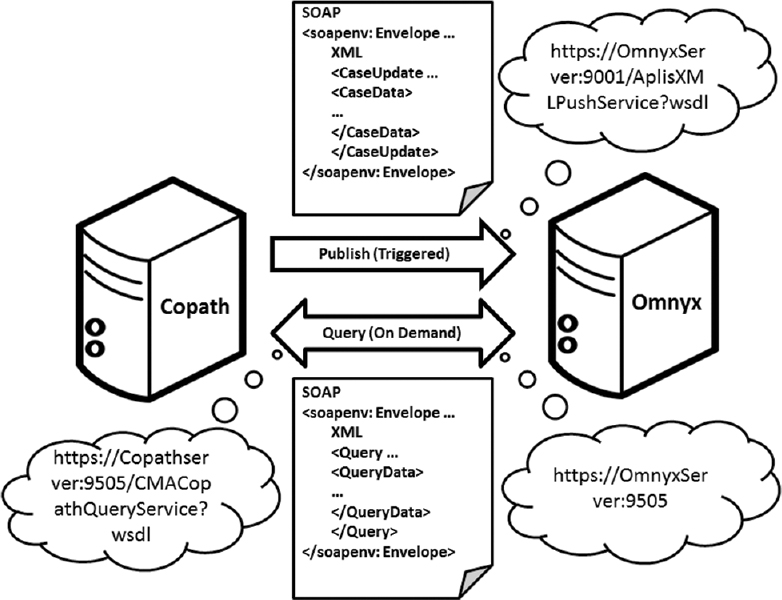
What happens when you integrate a digital pathology system (DPS) with an anatomical pathology laboratory information system (APLIS)? In the case of Guo et al. and the University of Pittsburgh Medical Center, "[t]he integration streamlined our digital sign-out workflow, diminished the potential for human error related to matching slides, and improved the sign-out experience for pathologists." This paper, published in
Journal of Pathology Informatics in 2016, describes their line of thinking, integration plans, and final results.
Posted on May 5, 2016
By John Jones
Journal articles
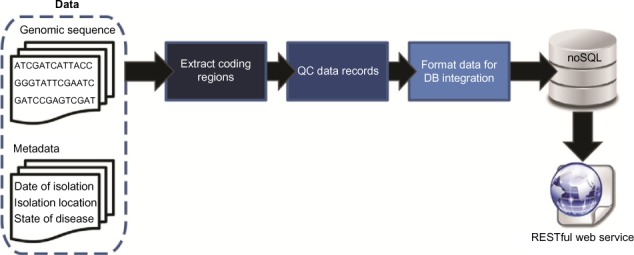
In this 2016 paper published in
Evolutionary Bioinformatics, Reisman
et al. discuss a polyglot approach "involving multiple languages, libraries, and persistence mechanisms" towards managing genomic sequence data. Using a NoSQL and RESTful web service approach, the team tested their developed pipeline on an evolutionary study of HIV-1. They conclude that " the case study highlights the abilities of the tool," and "although utilized for the investigation of a virus here, the approach can be applied to any species of interest."
Posted on April 25, 2016
By John Jones
Journal articles
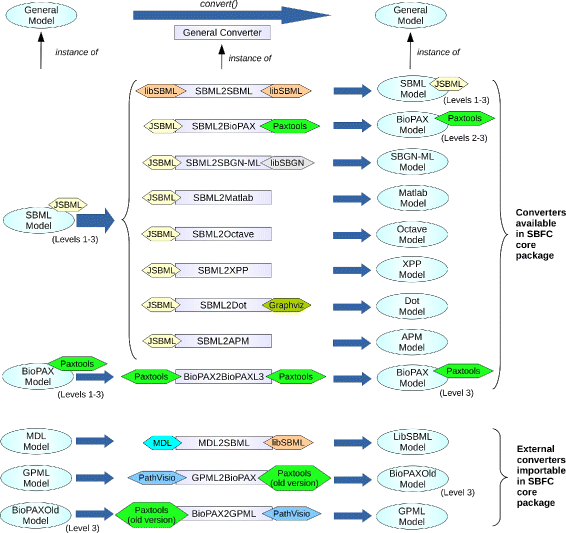
Rodriguez
et al. found that when converting computational models from one format to another, while many tools exist, they tend not to be very interoperable and can often be redundant. Additionally, they can be unmaintained or left abandoned. The researchers saw a need for a modular, open-source software system "to support rapid implementation and integration of new converters" in a more collaboratory way. They developed the System Biology Format Converter (SBFC), a Java-based tool that, per their conclusion, "helps computational biologists to process or visualise their models using different software tools, and software developers to implement format conversion."
Posted on April 19, 2016
By John Jones
Journal articles
In this 2015 journal article published in the open-access journal
Journal of Cheminformatics, Mohebifar and Sajadi describe their web-based HTML5/CSS3 3D molecule editor and visualizer Chemozart. Able to be run from the public web source or your own personal instance, Chemozart is both useful for educational and research purposes. The authors tout "that there’s no need to install anything and it can be accessed easily via a URL."
Posted on April 13, 2016
By John Jones
Journal articles
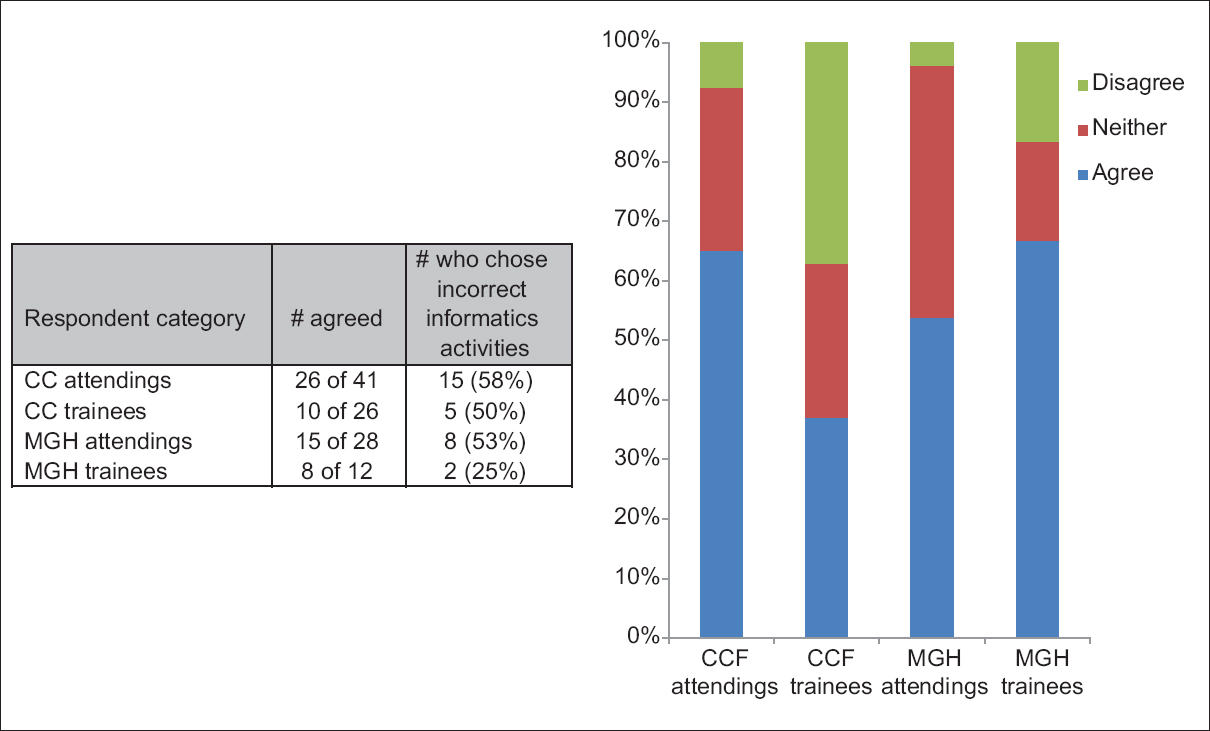
Perhaps frustrated with the state of education and misperceptions in regards to pathology informatics (PI), Walker
et al. set out to conduct a survey of noninformatics-oriented pathologists and trainees at the Cleveland Clinic and Massachusetts General Hospital to better grasp views of the professional field. In this paper published in
Journal of Pathology Informatics in April 2016, the researchers present their findings and opine about the state of pathology informatics education. They conclude: "Improved understanding and acceptance of PI throughout the pathology community could facilitate the communication and cooperation necessary to realize the type of informatics initiatives capable of advancing the importance of pathologists in the changing healthcare environment."
Posted on April 6, 2016
By John Jones
Journal articles

In this "opinion article" published in the open-access journal
F1000Research, Dirnagl and Przesdzing argue for the benefits of — and recognize the occasional problems with — electronic laboratory notebooks (ELNs). The duo attempts to answer the questions "What does it afford you, what does it require, and how should you go about implementing it?" They conclude that ELNs improve workflow and collaboration, allow for greater data integration, reduce transcription errors, improve data quality, and promote compliance with a variety of practices and regulations. "We have no doubt that ELNs will become standard in most life science laboratories in the near future," they state at the end.
Posted on April 2, 2016
By John Jones
Journal articles
Is digital diagnostic determination of pathology images quicker than microscopic? It depends on who you ask. Vodovnik published results of his comparative study in early 2016, comparing digital and microscopic diagnostic times using tools such as digital pathology workstations and laboratory information management systems. His conclusion: "A shorter diagnostic time in digital pathology comparing with traditional microscopy can be achieved in the routine diagnostic setting with adequate and stable network speeds, fully integrated LIMS and double displays as default parameters, in addition to better ergonomics, larger viewing field, and absence of physical slide handling, with effects on the both diagnostic and nondiagnostic time."
Posted on March 26, 2016
By John Jones
Journal articles
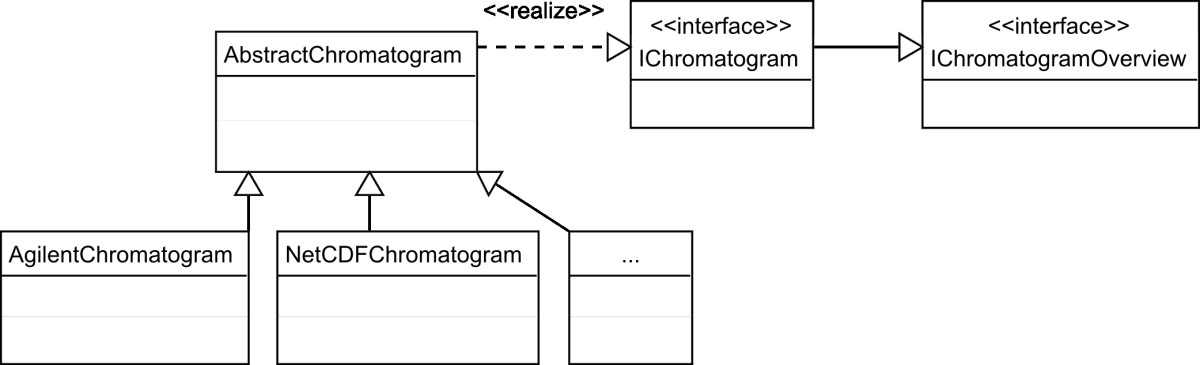
In 2010, Philip Wenig and Juergen Odermatt published their experiences on developing OpenChrom, an open-source chromatography data management system (CDMS) for analyzing mass spectrometric chromatographic data. The team built the software due to a perceived "lack of software systems that are capable to enhance nominal mass spectral data files, that are flexible, extensible and that offer an easy to use graphical user interface." The group concluded that "OpenChrom will be hopefully extended by contributing developers, scientists and companies in the future." As of 2016, development on OpenChrom does indeed continue, with a 1.1.0 preview release available released in February.
Posted on March 23, 2016
By John Jones
Journal articles

This 2012 paper, published in
Journal of Pathology Informatics, provides the perspective of Yale University School of Medicine's Pathology Informatics Unit in regards to their experience with custom software development. Despite perceptions concerning the pitfalls of laboratories developing their own software, Sinard
et al. conclude that "[m]any of the risks associated with custom development can be mitigated by a well-structured development process, use of open-source tools, and embracing an agile development philosophy."
Posted on March 15, 2016
By John Jones
Journal articles
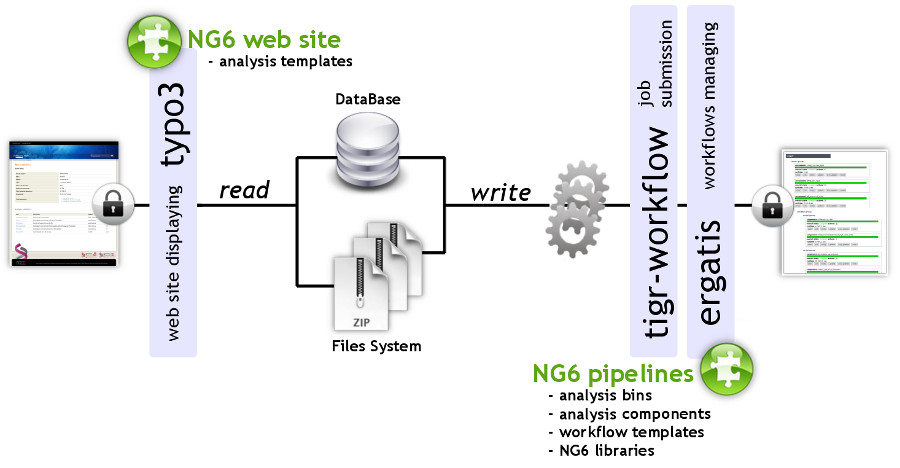
Published in 2012, this paper on the NG6 next generation sequencing (NGS) platform describes the software's development process at GenoToul, a French shared life sciences research facility. Mariette
et al. found that NGS software Galaxy was promising but not sufficient as it only "aims at simplifying data processing for researchers," whereas the group needed a LIMS that was also good at "gathering specialized pipelines and website" data. NG6 was developed to meet that need.
Posted on March 14, 2016
By John Jones
Journal articles

Hernández-de-Diego et al. from the Centro de Investigación Príncipe Felipe and the Karolinska Institute needed a softwar that " [i]n contrast to other solutions that put the focus on management of thousands of samples for core sequencing facilities ... [could handle] the annotation of experiments designed and run at individual research laboratories." Additionally, genomics solutions such as BASE had annotation limitations in regards to microarray experiment data. The group decided to develop their own system called the STATegra experiment management system (EMS), which they concluded "provides an integrated system for annotation of complex high-throughput omics experiments at functional genomics research laboratories."
Posted on March 3, 2016
By John Jones
Journal articles

In this 2012 journal article from
ZooKeys, Blagoderov and his colleagues from the Natural History Museum in London describe the process they went through to improve the mass-digitization of their numerous collections. Noting problems with cost, "as well as the inherent fragmentation in collection based biodiversity informatics," the researchers created a "wall-to-wall" process using the SatScan tray scanning system. The group suggested that such a system could, when implemented well, "open up collections to the world, facilitating their use, and help create a global collection index that can be used to set priorities for further digitization."
Posted on March 3, 2016
By John Jones
Journal articles
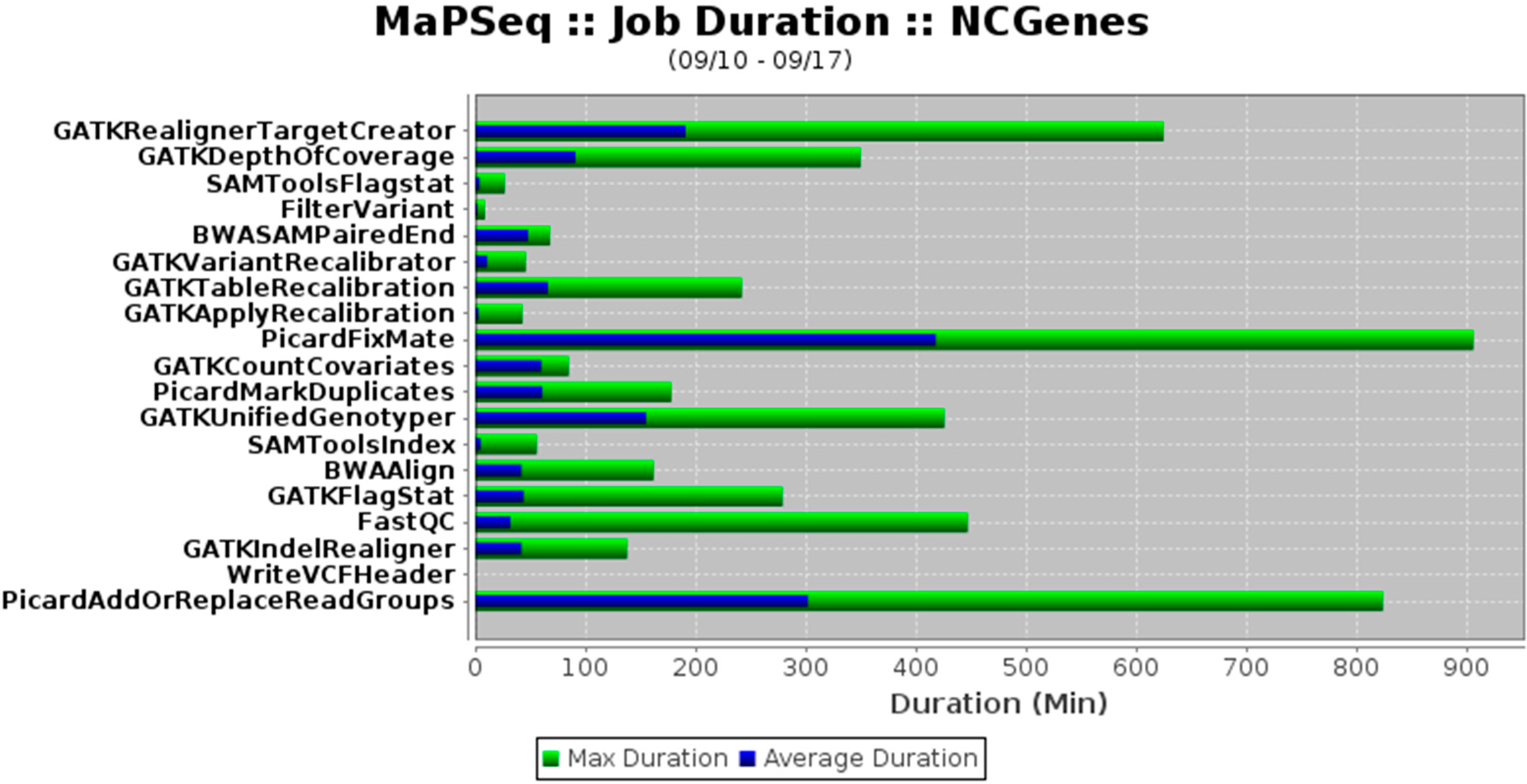
Reilly et al. at the University of North Carolina needed a specific solution for their genomics research, a solution that would "address the organizational challenges of computation-intensive biomedical research within a decentralized academic institution." After reviewing numerous options, the team decided to create their own system, MaPSeq, an application built on service-oriented architecture. In this paper summing up their development and implementation process, the group concludes that while the software is useful for massively parallel sequencing, "the general architecture and approach can be adapted for other complex or computationally-intense workflows."
Posted on February 15, 2016
By John Jones
Journal articles

In this brief article published in
Frontiers in Environmental Science in 2014, environmental researcher Alexander Kokhanovsky takes a look at the state of environmental informatics past and present while also speculating about the future challenges. He ends by emphasizing the need for improved "estimation of aerosol load using space-borne instrumentation" to improve environmental research, including improved algorithms and visualization tools.
Posted on February 12, 2016
By John Jones
Journal articles
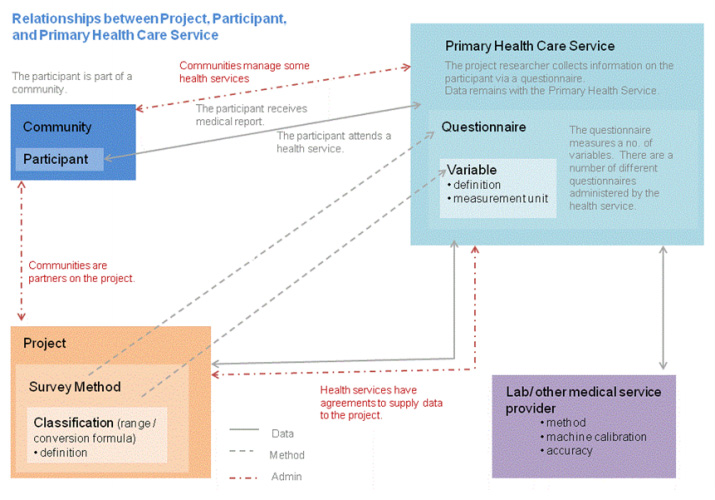
van Gaans
et al. of the University of South Australia and James Cook University needed a system that provides data management functionality for the specific needs of a public health research organization, including data sharing and reuse. They needed a system that would "ensure that: data that are unmanaged be managed, data that are disconnected be connected, data that are invisible be findable, [and] data that are single use be reusable, within a structured collection." In this journal article, the group documents the development of their answer: the Public Health Data Management System (PHRDMS).
Posted on January 28, 2016
By John Jones
Journal articles
Forensic science researcher Bruce Levy of the University of Illinois at Chicago presents his ideas about how forensic pathologists and clinical informaticians can address the historical and current shortcomings of putting sudden and violent death data to better use. Levy argues that improved collaborations and data standards "will enable forensic pathology to maximize its effectiveness by providing timely and actionable information to public health and public safety agencies."
Posted on January 21, 2016
By John Jones
Journal articles
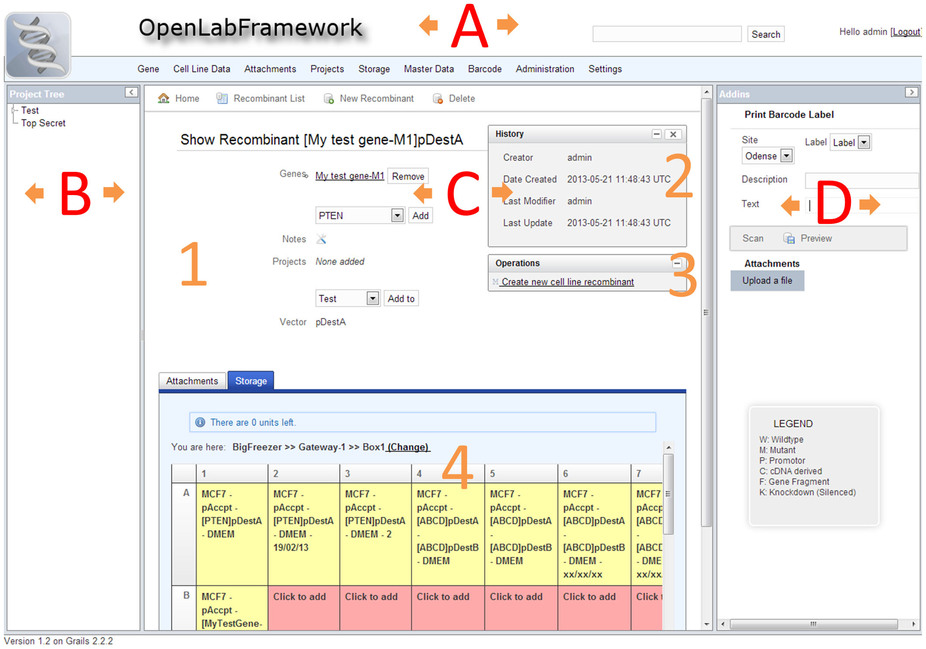
Nanomedicine researchers at the University of Southern Denmark required a "dedicated LIMS for the management of large vector construct and cell line libraries." With no other open-source options available, the team developed their own, OpenLabFramework. Documenting their process, List
et al. conclude "OLF can be deployed using different database management systems either locally, to a server, or to the cloud," and "[t]he incorporation of modern technologies, such as mobile devices and printing of barcode labels may increase productivity even further."
Posted on January 12, 2016
By John Jones
Journal articles
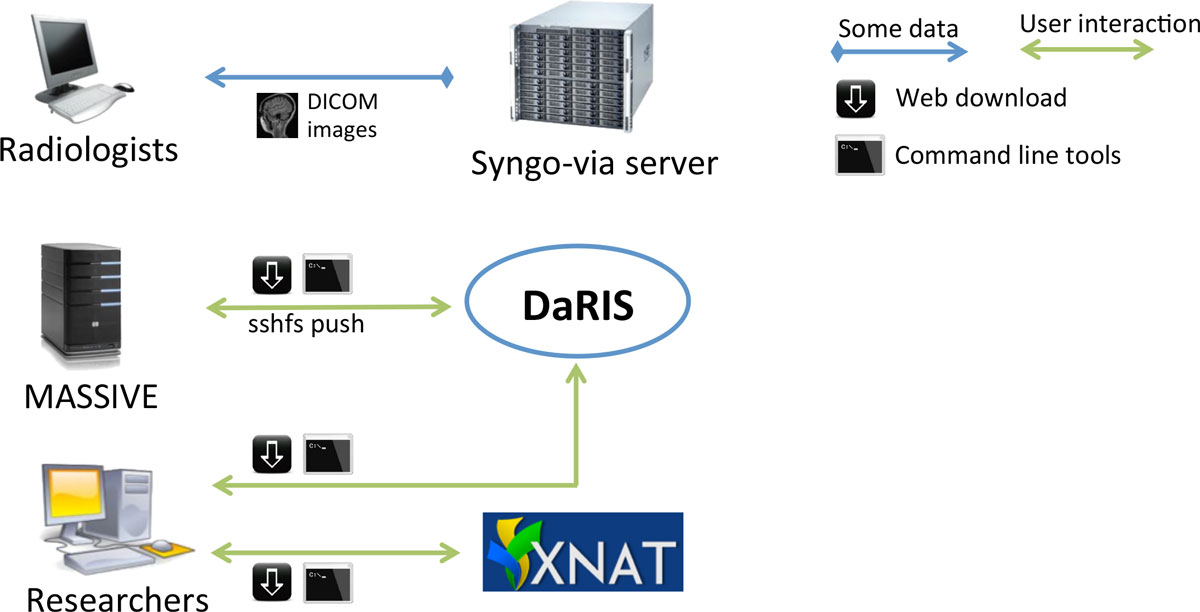
In this research paper, Nguyen et al. of Monash University in Australia "describes the design, implementation and operation of a multi-modality research imaging data management system that manages imaging data obtained from biomedical imaging scanners" at their facility. Faced with limitations of existing image management software and frameworks, the group custom built a system "based on DaRIS and XNAT has been designed and implemented to enable researchers to acquire, manage and analyse large, longitudinal biomedical imaging datasets."
Posted on January 11, 2016
By John Jones
Journal articles
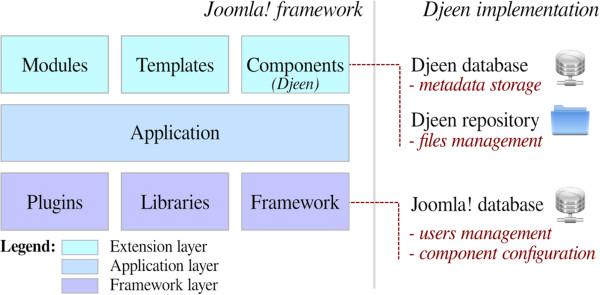
Stahl et al. and Universités de Montpellier needed a system that could "streamline [biological] data storage and annotation collaboratively." Not finding a system to their liking, the group developed a Joomla!-based LIMS called Djeen and published their research on the development process in 2013. The group concludes: "Djeen allows managing project associated with heterogeneous data types while enforcing annotation integrity and minimum information. Projects are managed within a hierarchy and user permissions are finely-grained for each project, user and group."
 The state of data management across the sciences is getting increasingly complex as data stores build up, and the world of public health is no less affected. Making sense of data is one portion of management, but quality analysis is also an important but slightly understated aspect as well. This 2015 paper by Eaton et al. explains a series of "data quality tools developed to gain insight into the data quality problems associated with these data." The group concludes "our key insight was the need to assess temporal patterns in the data in terms of accrual lag."
The state of data management across the sciences is getting increasingly complex as data stores build up, and the world of public health is no less affected. Making sense of data is one portion of management, but quality analysis is also an important but slightly understated aspect as well. This 2015 paper by Eaton et al. explains a series of "data quality tools developed to gain insight into the data quality problems associated with these data." The group concludes "our key insight was the need to assess temporal patterns in the data in terms of accrual lag."
 What happens when you integrate a digital pathology system (DPS) with an anatomical pathology laboratory information system (APLIS)? In the case of Guo et al. and the University of Pittsburgh Medical Center, "[t]he integration streamlined our digital sign-out workflow, diminished the potential for human error related to matching slides, and improved the sign-out experience for pathologists." This paper, published in Journal of Pathology Informatics in 2016, describes their line of thinking, integration plans, and final results.
What happens when you integrate a digital pathology system (DPS) with an anatomical pathology laboratory information system (APLIS)? In the case of Guo et al. and the University of Pittsburgh Medical Center, "[t]he integration streamlined our digital sign-out workflow, diminished the potential for human error related to matching slides, and improved the sign-out experience for pathologists." This paper, published in Journal of Pathology Informatics in 2016, describes their line of thinking, integration plans, and final results.
 In this 2016 paper published in Evolutionary Bioinformatics, Reisman et al. discuss a polyglot approach "involving multiple languages, libraries, and persistence mechanisms" towards managing genomic sequence data. Using a NoSQL and RESTful web service approach, the team tested their developed pipeline on an evolutionary study of HIV-1. They conclude that " the case study highlights the abilities of the tool," and "although utilized for the investigation of a virus here, the approach can be applied to any species of interest."
In this 2016 paper published in Evolutionary Bioinformatics, Reisman et al. discuss a polyglot approach "involving multiple languages, libraries, and persistence mechanisms" towards managing genomic sequence data. Using a NoSQL and RESTful web service approach, the team tested their developed pipeline on an evolutionary study of HIV-1. They conclude that " the case study highlights the abilities of the tool," and "although utilized for the investigation of a virus here, the approach can be applied to any species of interest."
 Rodriguez et al. found that when converting computational models from one format to another, while many tools exist, they tend not to be very interoperable and can often be redundant. Additionally, they can be unmaintained or left abandoned. The researchers saw a need for a modular, open-source software system "to support rapid implementation and integration of new converters" in a more collaboratory way. They developed the System Biology Format Converter (SBFC), a Java-based tool that, per their conclusion, "helps computational biologists to process or visualise their models using different software tools, and software developers to implement format conversion."
Rodriguez et al. found that when converting computational models from one format to another, while many tools exist, they tend not to be very interoperable and can often be redundant. Additionally, they can be unmaintained or left abandoned. The researchers saw a need for a modular, open-source software system "to support rapid implementation and integration of new converters" in a more collaboratory way. They developed the System Biology Format Converter (SBFC), a Java-based tool that, per their conclusion, "helps computational biologists to process or visualise their models using different software tools, and software developers to implement format conversion."
 In this 2015 journal article published in the open-access journal Journal of Cheminformatics, Mohebifar and Sajadi describe their web-based HTML5/CSS3 3D molecule editor and visualizer Chemozart. Able to be run from the public web source or your own personal instance, Chemozart is both useful for educational and research purposes. The authors tout "that there’s no need to install anything and it can be accessed easily via a URL."
In this 2015 journal article published in the open-access journal Journal of Cheminformatics, Mohebifar and Sajadi describe their web-based HTML5/CSS3 3D molecule editor and visualizer Chemozart. Able to be run from the public web source or your own personal instance, Chemozart is both useful for educational and research purposes. The authors tout "that there’s no need to install anything and it can be accessed easily via a URL."
 Perhaps frustrated with the state of education and misperceptions in regards to pathology informatics (PI), Walker et al. set out to conduct a survey of noninformatics-oriented pathologists and trainees at the Cleveland Clinic and Massachusetts General Hospital to better grasp views of the professional field. In this paper published in Journal of Pathology Informatics in April 2016, the researchers present their findings and opine about the state of pathology informatics education. They conclude: "Improved understanding and acceptance of PI throughout the pathology community could facilitate the communication and cooperation necessary to realize the type of informatics initiatives capable of advancing the importance of pathologists in the changing healthcare environment."
Perhaps frustrated with the state of education and misperceptions in regards to pathology informatics (PI), Walker et al. set out to conduct a survey of noninformatics-oriented pathologists and trainees at the Cleveland Clinic and Massachusetts General Hospital to better grasp views of the professional field. In this paper published in Journal of Pathology Informatics in April 2016, the researchers present their findings and opine about the state of pathology informatics education. They conclude: "Improved understanding and acceptance of PI throughout the pathology community could facilitate the communication and cooperation necessary to realize the type of informatics initiatives capable of advancing the importance of pathologists in the changing healthcare environment."
 In this "opinion article" published in the open-access journal F1000Research, Dirnagl and Przesdzing argue for the benefits of — and recognize the occasional problems with — electronic laboratory notebooks (ELNs). The duo attempts to answer the questions "What does it afford you, what does it require, and how should you go about implementing it?" They conclude that ELNs improve workflow and collaboration, allow for greater data integration, reduce transcription errors, improve data quality, and promote compliance with a variety of practices and regulations. "We have no doubt that ELNs will become standard in most life science laboratories in the near future," they state at the end.
In this "opinion article" published in the open-access journal F1000Research, Dirnagl and Przesdzing argue for the benefits of — and recognize the occasional problems with — electronic laboratory notebooks (ELNs). The duo attempts to answer the questions "What does it afford you, what does it require, and how should you go about implementing it?" They conclude that ELNs improve workflow and collaboration, allow for greater data integration, reduce transcription errors, improve data quality, and promote compliance with a variety of practices and regulations. "We have no doubt that ELNs will become standard in most life science laboratories in the near future," they state at the end.
 Is digital diagnostic determination of pathology images quicker than microscopic? It depends on who you ask. Vodovnik published results of his comparative study in early 2016, comparing digital and microscopic diagnostic times using tools such as digital pathology workstations and laboratory information management systems. His conclusion: "A shorter diagnostic time in digital pathology comparing with traditional microscopy can be achieved in the routine diagnostic setting with adequate and stable network speeds, fully integrated LIMS and double displays as default parameters, in addition to better ergonomics, larger viewing field, and absence of physical slide handling, with effects on the both diagnostic and nondiagnostic time."
Is digital diagnostic determination of pathology images quicker than microscopic? It depends on who you ask. Vodovnik published results of his comparative study in early 2016, comparing digital and microscopic diagnostic times using tools such as digital pathology workstations and laboratory information management systems. His conclusion: "A shorter diagnostic time in digital pathology comparing with traditional microscopy can be achieved in the routine diagnostic setting with adequate and stable network speeds, fully integrated LIMS and double displays as default parameters, in addition to better ergonomics, larger viewing field, and absence of physical slide handling, with effects on the both diagnostic and nondiagnostic time."
 In 2010, Philip Wenig and Juergen Odermatt published their experiences on developing OpenChrom, an open-source chromatography data management system (CDMS) for analyzing mass spectrometric chromatographic data. The team built the software due to a perceived "lack of software systems that are capable to enhance nominal mass spectral data files, that are flexible, extensible and that offer an easy to use graphical user interface." The group concluded that "OpenChrom will be hopefully extended by contributing developers, scientists and companies in the future." As of 2016, development on OpenChrom does indeed continue, with a 1.1.0 preview release available released in February.
In 2010, Philip Wenig and Juergen Odermatt published their experiences on developing OpenChrom, an open-source chromatography data management system (CDMS) for analyzing mass spectrometric chromatographic data. The team built the software due to a perceived "lack of software systems that are capable to enhance nominal mass spectral data files, that are flexible, extensible and that offer an easy to use graphical user interface." The group concluded that "OpenChrom will be hopefully extended by contributing developers, scientists and companies in the future." As of 2016, development on OpenChrom does indeed continue, with a 1.1.0 preview release available released in February.
 This 2012 paper, published in Journal of Pathology Informatics, provides the perspective of Yale University School of Medicine's Pathology Informatics Unit in regards to their experience with custom software development. Despite perceptions concerning the pitfalls of laboratories developing their own software, Sinard et al. conclude that "[m]any of the risks associated with custom development can be mitigated by a well-structured development process, use of open-source tools, and embracing an agile development philosophy."
This 2012 paper, published in Journal of Pathology Informatics, provides the perspective of Yale University School of Medicine's Pathology Informatics Unit in regards to their experience with custom software development. Despite perceptions concerning the pitfalls of laboratories developing their own software, Sinard et al. conclude that "[m]any of the risks associated with custom development can be mitigated by a well-structured development process, use of open-source tools, and embracing an agile development philosophy."
 Published in 2012, this paper on the NG6 next generation sequencing (NGS) platform describes the software's development process at GenoToul, a French shared life sciences research facility. Mariette et al. found that NGS software Galaxy was promising but not sufficient as it only "aims at simplifying data processing for researchers," whereas the group needed a LIMS that was also good at "gathering specialized pipelines and website" data. NG6 was developed to meet that need.
Published in 2012, this paper on the NG6 next generation sequencing (NGS) platform describes the software's development process at GenoToul, a French shared life sciences research facility. Mariette et al. found that NGS software Galaxy was promising but not sufficient as it only "aims at simplifying data processing for researchers," whereas the group needed a LIMS that was also good at "gathering specialized pipelines and website" data. NG6 was developed to meet that need.
 Hernández-de-Diego et al. from the Centro de Investigación Príncipe Felipe and the Karolinska Institute needed a softwar that " [i]n contrast to other solutions that put the focus on management of thousands of samples for core sequencing facilities ... [could handle] the annotation of experiments designed and run at individual research laboratories." Additionally, genomics solutions such as BASE had annotation limitations in regards to microarray experiment data. The group decided to develop their own system called the STATegra experiment management system (EMS), which they concluded "provides an integrated system for annotation of complex high-throughput omics experiments at functional genomics research laboratories."
Hernández-de-Diego et al. from the Centro de Investigación Príncipe Felipe and the Karolinska Institute needed a softwar that " [i]n contrast to other solutions that put the focus on management of thousands of samples for core sequencing facilities ... [could handle] the annotation of experiments designed and run at individual research laboratories." Additionally, genomics solutions such as BASE had annotation limitations in regards to microarray experiment data. The group decided to develop their own system called the STATegra experiment management system (EMS), which they concluded "provides an integrated system for annotation of complex high-throughput omics experiments at functional genomics research laboratories."
 In this 2012 journal article from ZooKeys, Blagoderov and his colleagues from the Natural History Museum in London describe the process they went through to improve the mass-digitization of their numerous collections. Noting problems with cost, "as well as the inherent fragmentation in collection based biodiversity informatics," the researchers created a "wall-to-wall" process using the SatScan tray scanning system. The group suggested that such a system could, when implemented well, "open up collections to the world, facilitating their use, and help create a global collection index that can be used to set priorities for further digitization."
In this 2012 journal article from ZooKeys, Blagoderov and his colleagues from the Natural History Museum in London describe the process they went through to improve the mass-digitization of their numerous collections. Noting problems with cost, "as well as the inherent fragmentation in collection based biodiversity informatics," the researchers created a "wall-to-wall" process using the SatScan tray scanning system. The group suggested that such a system could, when implemented well, "open up collections to the world, facilitating their use, and help create a global collection index that can be used to set priorities for further digitization."
 Reilly et al. at the University of North Carolina needed a specific solution for their genomics research, a solution that would "address the organizational challenges of computation-intensive biomedical research within a decentralized academic institution." After reviewing numerous options, the team decided to create their own system, MaPSeq, an application built on service-oriented architecture. In this paper summing up their development and implementation process, the group concludes that while the software is useful for massively parallel sequencing, "the general architecture and approach can be adapted for other complex or computationally-intense workflows."
Reilly et al. at the University of North Carolina needed a specific solution for their genomics research, a solution that would "address the organizational challenges of computation-intensive biomedical research within a decentralized academic institution." After reviewing numerous options, the team decided to create their own system, MaPSeq, an application built on service-oriented architecture. In this paper summing up their development and implementation process, the group concludes that while the software is useful for massively parallel sequencing, "the general architecture and approach can be adapted for other complex or computationally-intense workflows."
 In this brief article published in Frontiers in Environmental Science in 2014, environmental researcher Alexander Kokhanovsky takes a look at the state of environmental informatics past and present while also speculating about the future challenges. He ends by emphasizing the need for improved "estimation of aerosol load using space-borne instrumentation" to improve environmental research, including improved algorithms and visualization tools.
In this brief article published in Frontiers in Environmental Science in 2014, environmental researcher Alexander Kokhanovsky takes a look at the state of environmental informatics past and present while also speculating about the future challenges. He ends by emphasizing the need for improved "estimation of aerosol load using space-borne instrumentation" to improve environmental research, including improved algorithms and visualization tools.
 van Gaans et al. of the University of South Australia and James Cook University needed a system that provides data management functionality for the specific needs of a public health research organization, including data sharing and reuse. They needed a system that would "ensure that: data that are unmanaged be managed, data that are disconnected be connected, data that are invisible be findable, [and] data that are single use be reusable, within a structured collection." In this journal article, the group documents the development of their answer: the Public Health Data Management System (PHRDMS).
van Gaans et al. of the University of South Australia and James Cook University needed a system that provides data management functionality for the specific needs of a public health research organization, including data sharing and reuse. They needed a system that would "ensure that: data that are unmanaged be managed, data that are disconnected be connected, data that are invisible be findable, [and] data that are single use be reusable, within a structured collection." In this journal article, the group documents the development of their answer: the Public Health Data Management System (PHRDMS).
 Nanomedicine researchers at the University of Southern Denmark required a "dedicated LIMS for the management of large vector construct and cell line libraries." With no other open-source options available, the team developed their own, OpenLabFramework. Documenting their process, List et al. conclude "OLF can be deployed using different database management systems either locally, to a server, or to the cloud," and "[t]he incorporation of modern technologies, such as mobile devices and printing of barcode labels may increase productivity even further."
Nanomedicine researchers at the University of Southern Denmark required a "dedicated LIMS for the management of large vector construct and cell line libraries." With no other open-source options available, the team developed their own, OpenLabFramework. Documenting their process, List et al. conclude "OLF can be deployed using different database management systems either locally, to a server, or to the cloud," and "[t]he incorporation of modern technologies, such as mobile devices and printing of barcode labels may increase productivity even further."
 In this research paper, Nguyen et al. of Monash University in Australia "describes the design, implementation and operation of a multi-modality research imaging data management system that manages imaging data obtained from biomedical imaging scanners" at their facility. Faced with limitations of existing image management software and frameworks, the group custom built a system "based on DaRIS and XNAT has been designed and implemented to enable researchers to acquire, manage and analyse large, longitudinal biomedical imaging datasets."
In this research paper, Nguyen et al. of Monash University in Australia "describes the design, implementation and operation of a multi-modality research imaging data management system that manages imaging data obtained from biomedical imaging scanners" at their facility. Faced with limitations of existing image management software and frameworks, the group custom built a system "based on DaRIS and XNAT has been designed and implemented to enable researchers to acquire, manage and analyse large, longitudinal biomedical imaging datasets."
 Stahl et al. and Universités de Montpellier needed a system that could "streamline [biological] data storage and annotation collaboratively." Not finding a system to their liking, the group developed a Joomla!-based LIMS called Djeen and published their research on the development process in 2013. The group concludes: "Djeen allows managing project associated with heterogeneous data types while enforcing annotation integrity and minimum information. Projects are managed within a hierarchy and user permissions are finely-grained for each project, user and group."
Stahl et al. and Universités de Montpellier needed a system that could "streamline [biological] data storage and annotation collaboratively." Not finding a system to their liking, the group developed a Joomla!-based LIMS called Djeen and published their research on the development process in 2013. The group concludes: "Djeen allows managing project associated with heterogeneous data types while enforcing annotation integrity and minimum information. Projects are managed within a hierarchy and user permissions are finely-grained for each project, user and group."















