Posted on February 27, 2018
By Shawn Douglas
Journal articles
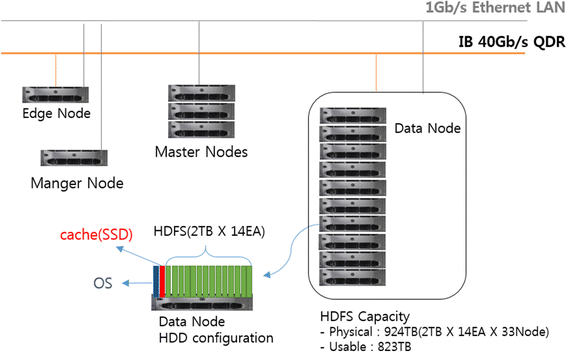
Ko
et al.of the Korean BioInformation Center discuss their Closha web service for large-scale genomic data analysis in this 2018 paper published in
BMC Bioinformatics. Noting a lack of rapid, cost-effective genomics workflow capable of running all the major genomic data analysis application in one pipeline, the researchers developed a hybrid system that can combine workflows. Additionally, they developed an add-on tool that handles the sheer size of genomic files and the speed with which they transfer, reaching transfer speeds "of up to 10 times that of normal FTP and HTTP." They conclude that "Closha allows genomic researchers without informatics or programming expertise to perform complex large-scale analysis with only a web browser."
Posted on February 20, 2018
By Shawn Douglas
Journal articles
In this 2018 paper published in
Scientific Programming, Zhu
et al. review the state of big data in the geological sciences and provide context to the challenges associated with managing that data in the cloud using China's various databases and tools as examples. Using the term "cloud-enabled geological information services" or CEGIS, they also outline the existing and new technologies that will bring together and shape how geologic data is accessed and used in the cloud. They conclude that "[w]ith the continuous development of big data technologies in addressing those challenges related to geological big data, such as the difficulties of describing and modeling geological big data with some complex characteristics, CEGIS will move towards a more mature and more intelligent direction in the future."
Posted on February 15, 2018
By Shawn Douglas
Journal articles
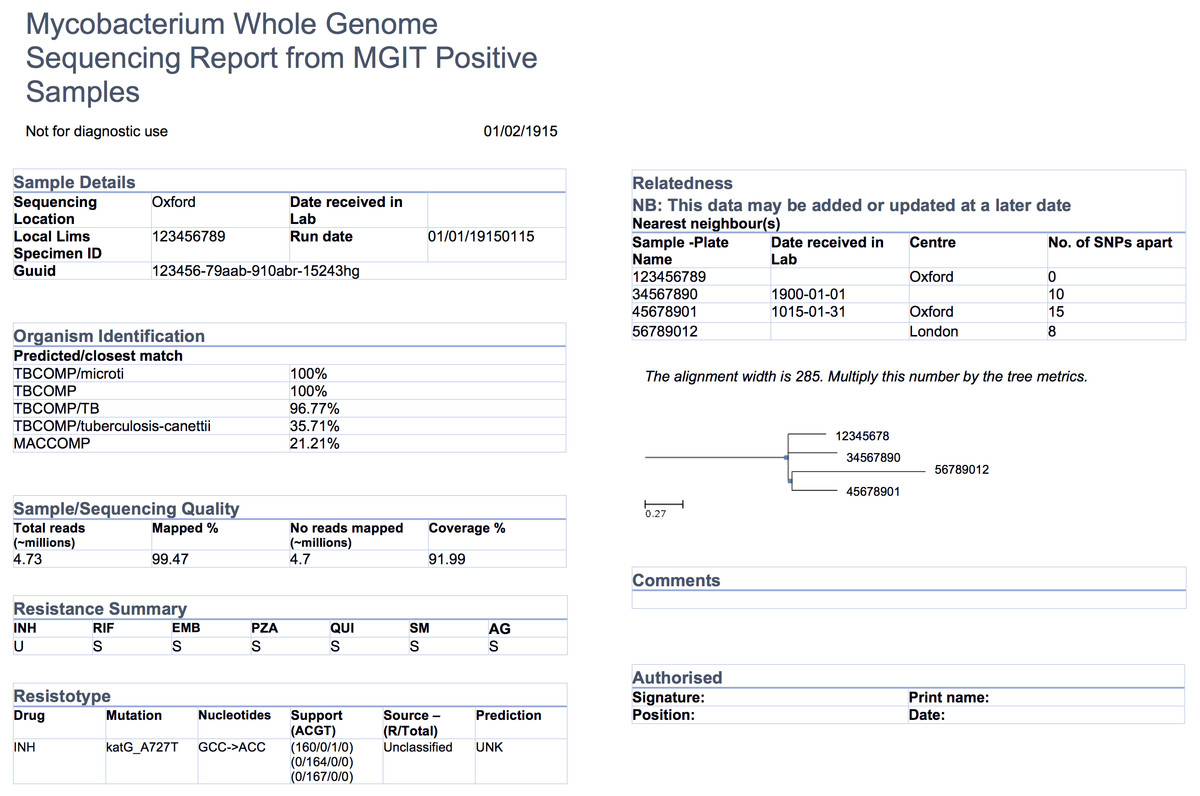
Reporting isn't as simple as casually placing key figures on a page; significant work should go into designing a report template, particularly those reporting specialized data , like that found in the world of pathogen genomics. Crisan
et al. of the University of British Columbia and the BC Centre for Disease Control looked for evidence-based guidelines on creating reports for such a specialty — specifically for tuberculosis genomic testing — and couldn't find any. So they researched and created their own. This 2018 paper details their journey towards a final report design, concluding "that the application of human-centered design methodologies allowed us to improve not only the visual aesthetics of the final report, but also its functionality, by carefully coupling stakeholder tasks, data, and constraints to techniques from information and graphic design."
Posted on February 6, 2018
By Shawn Douglas
Journal articles
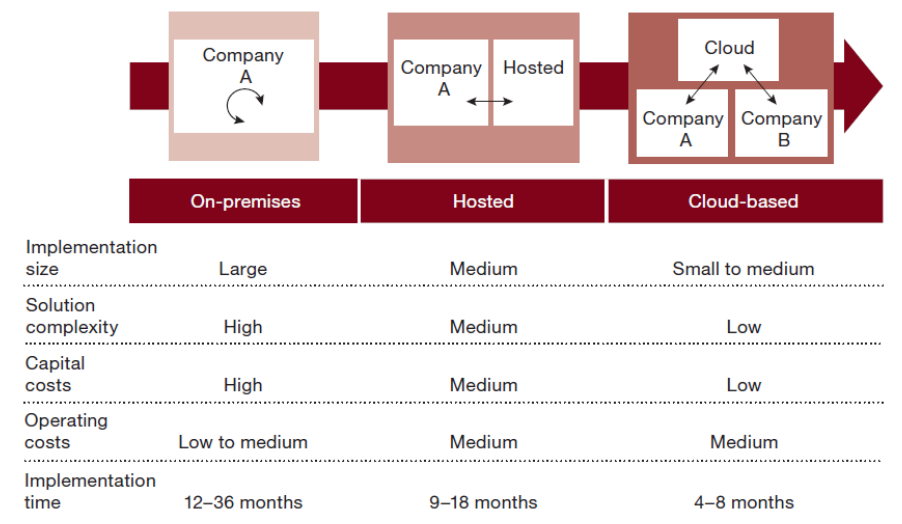
This 2017 paper by Saa
et al. examines the existing research literature concerning the status of moving an enterprise resource planning (ERP) system to the cloud. Noting both the many benefits of cloud for ERP and the data security drawbacks, the researchers discover the use of hybrid cloud-based ERPs by some organizations as a way to strike a balance, enabling them to "benefit from the agility and scalability of cloud-based ERP solutions while still keeping the security advantages from on-premise solutions for their mission-critical data."
Posted on January 30, 2018
By Shawn Douglas
Journal articles

A major goal among researchers in the age of big data is to improve data sharing and collaborative efforts. Kindler
et al. note, for example, that "specialized techniques for the production and analysis of viral metagenomes remains in a subset of labs," and other researchers likely don't know about them. As such, the authors of this 2017 paper in
F1000Research discuss the enhancement of the protocols.io online collaborative space to include more social elements "for scientists to share improvements and corrections to protocols so that others are not continuously re-discovering knowledge that scientists have not had the time or wear-with-all to publish." With its many connection points, they conclude, the software update "will allow the forum to evolve naturally given rapidly developing trends and new protocols."
Posted on January 23, 2018
By Shawn Douglas
Journal articles
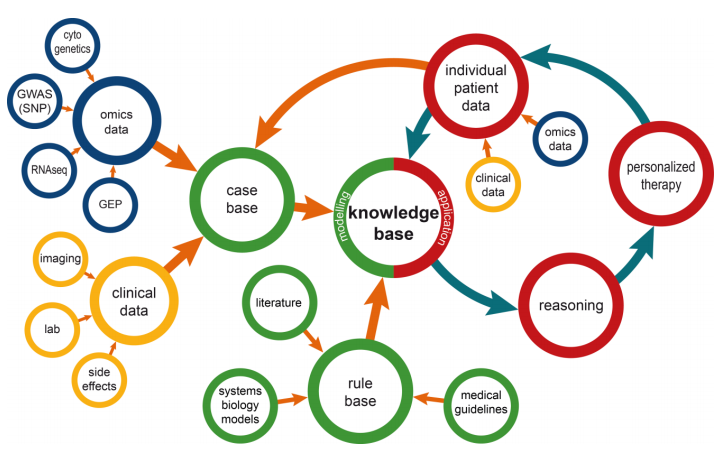
This brief paper by Ganzinger and Knaup examines the state of systems medicine, where multiple medical data streams are merged, analyzed, modeled, etc. to further how we diagnose and treat disease. They discuss the dynamic nature of disease knowledge and clinical data, as well as the problems that arise from integrating omics data into systems medicine, under the umbrella of integrating the knowledge into a usable format in tools like a clinical decision support system. They conclude that though with many benefits, "special care has to be taken to address inherent dynamics of data that are used for systems medicine; over time the number of available health records will increase and treatment approaches will change." They add that their data management model is flexible and can be used with other data management tools.
Posted on January 16, 2018
By Shawn Douglas
Journal articles
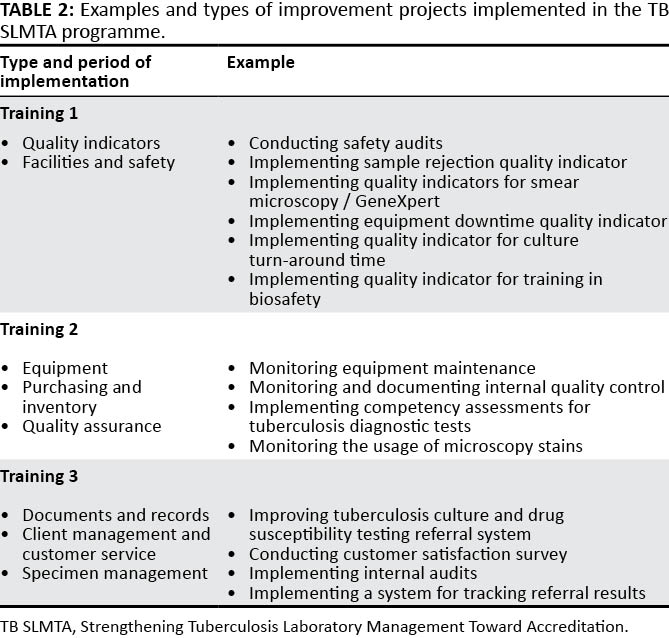
Published in early 2017, this paper by Albert
et al. discusses the development process of an accreditation program — the Strengthening Tuberculosis Laboratory Management Toward Accreditation or TB SLMTA — dedicated to better implementation of quality management systems (QMS) in tuberculosis laboratories around the world. The authors discuss the development of the curriculum, accreditation tools, and roll-out across 10 countries and 37 laboratories. They conclude that the training and mentoring program is increasingly vital for tuberculosis labs, "building a foundation toward further quality improvement toward achieving accreditation ... on the African continent and beyond."
Posted on January 8, 2018
By Shawn Douglas
Journal articles
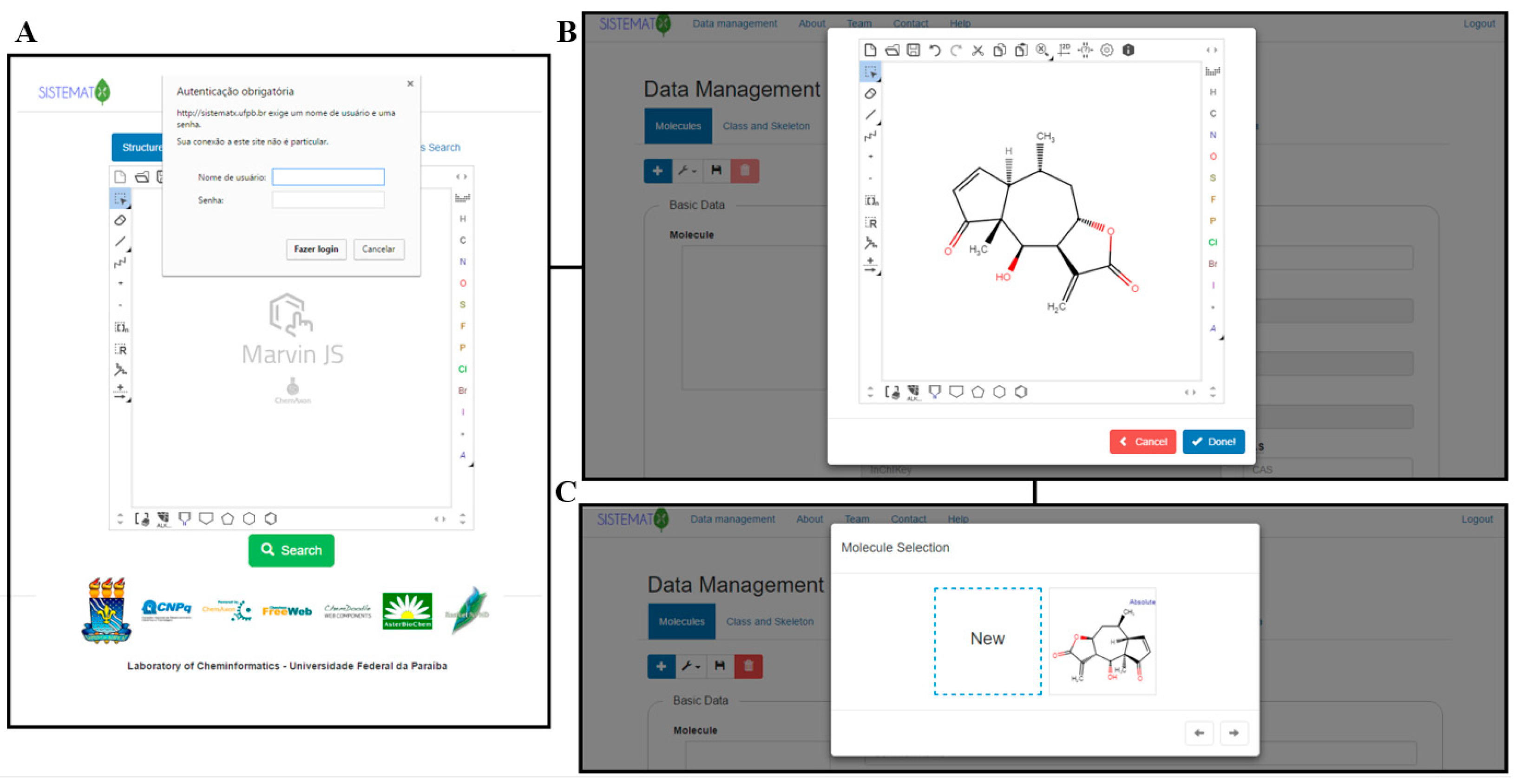
Dereplication is a chemical screening process of separation and purification that eliminates known and studied constituents, leaving other novel metabolites for future study. This is an important part of pharmaceutical and natural product development, requiring appropriate data management and retrieval for researchers around the world. However, Scotti
et al. found a lack of secondary metabolite databases that met their needs. Noting a need for complex searches, structure management, visualizations, and taxonomic rank in one package, they developed SistematX, "a modern and innovative web interface." They conclude the end result is a system that "provides a wealth of useful information for the scientific community about natural products, highlighting the location of species from which the compounds were isolated."
Posted on December 28, 2017
By Shawn Douglas
Journal articles
Social science researchers, like any other researchers, produce data in their work, both quantitative and qualitative. Unlike other sciences, social science researchers have some added difficulties in meeting the data sharing initiatives promoted by funding agencies and journals when it comes to qualitative data and how to share it "both ethically and safely." In this 2017 paper, Kirilova and Karcher describe the efforts of the Qualitative Data Repository at Syracuse University, considering the variables that went into making qualitative data available while trying to reconcile "the tension between modifying data or restricting access to them, and retaining their analytic value." They conclude that such repositories' goals should be towards "educating researchers how to be 'safe people' and how to plan for 'safe projects' – when accessing such data and using them for secondary analysis – and providing long-term 'safe settings' for the data, including via de-identification and appropriate access controls."
Posted on December 22, 2017
By Shawn Douglas
Journal articles
Metadata capture is important to scientific reproducibility argue Zehl
et al., particularly in regards to "the experiment, the data, and the applied preprocessing steps on the data." In this 2016 paper, however, the researchers demonstrate that metadata capture is not necessarily a simple task for an experimental laboratory; diverse datasets, specialized workflows, and lack of knowledge of supporting software tools all make the challenge of metadata capture more difficult. After demonstrating use cases representative of their neurophysiology lab and make a case for the open metadata Markup Language (odML), the researchers conclude that "[r]eadily available tools to support metadata management, such as odML, are a vital component in constructing [data and metadata management] workflows" in laboratories of all types.
Posted on December 12, 2017
By Shawn Douglas
Journal articles
In this 2017 paper by Plebani and Sciacovelli of the University Hospital of Padova, the duo offer their insights into the benefits and challenges of a clinical laboratory getting ISO 15189 accredited. Noting that in the European theater "major differences affect the approaches to accreditation promoted by the national bodies," the authors discuss the quality management approach that ISO 15189 prescribes and why its worth following. The conclude that while laboratories can realize "world-class quality and the need for a rigorous process of quality assurance," it still requires a high level of awareness among staff of the importance of ISO 15189 accreditation, an internal assessment plan, and well-defined, "suitable and user-friendly operating procedures."
Posted on December 4, 2017
By Shawn Douglas
Journal articles

In this 2017 paper by Curtin University's Cameron Neylon, the concepts of research data management (RDM) and research data sharing (RDS) are solidified and identified as increasingly common practices for funded research enterprises, particularly public-facing ones. But what of the implementation of these practices and the challenges associated with them? Neylon finds more than expected in his study, identifying sharp contrast among "those who saw [data management] requirements as a necessary part of raising issues among researchers and those concerned that this was leading to a compliance culture, where data management was seen as a required administrative exercise rather than an integral part of research practice." Neylon concludes with three key recommendations for researchers in general should follow to further shape how RDM and RDS are approached in research communities.
Posted on November 27, 2017
By Shawn Douglas
Journal articles
This brief review article from early 2017 looks at the basic elements of public health informatics and addresses how they're implemented. Aziz compares paper-based surveillance systems with electronic systems, noting the various improvements and challenges that have come with transitioning to electronic surveillance. He also reviews how public health informatics is applied in the U.S. and other parts of the world, including Saudi Arabia. Aziz concludes that "[p]atients, healthcare professionals, and public health officials can all help in reshaping public health through the adoption of new information systems, the use of electronic methods for disease surveillance, and the reformation of outmoded processes."
Posted on November 21, 2017
By Shawn Douglas
Journal articles

In this technical note from the University of Iowa Hospitals and Clinics, Imborek
et al. walk through the benefits and challenges of evaluating, implementing, and assessing preferred name and gender identity into its various laboratory systems and workflows. As preferred name has been an important point "in providing inclusion toward a class of patients that have historically been disenfranchised from the healthcare system" — primarily transgender patients — awareness is increasing at many levels about not only the inclusion of preferred name but also the complications that arise from it. From customizing software to billing and coding issues, from blood donor issues to laboratory test challenges, the group concludes that despite the "major undertaking [that] required the combined effort of many people from different teams," the efforts provide new opportunities for the field of pathology.
Posted on November 15, 2017
By Shawn Douglas
Journal articles
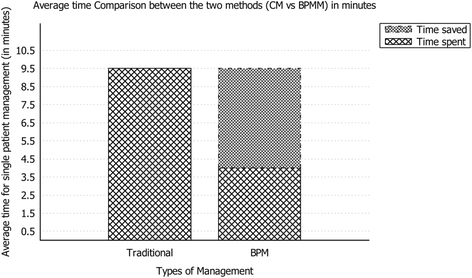
Sure, there's plenty of talk about big data in the research and clinical realms, but what of smarter data, gained from using business process management (BPM) tools? That's the direction Andellini
et al. went in after looking at the kidney transplantation cases at Bambino Gesù Children’s Hospital, realizing the potential for improving clinical decisions, scheduling of patient appointments, and reducing human errors in workflow. The group concluded "that the usage of BPM in the management of kidney transplantation leads to real benefits in terms of resource optimization and quality improvement," and that a "combination of a BPM-based platform with a service-oriented architecture could represent a revolution in clinical pathway management."
Posted on November 7, 2017
By Shawn Douglas
Journal articles
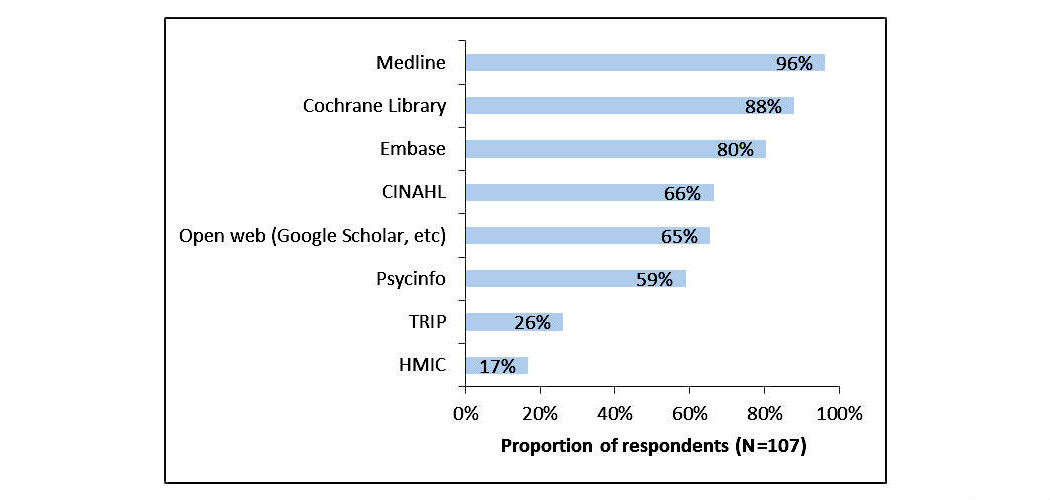
In this 2017 article published in
JMIR Medical Informatics, Russell-Rose and Chamberlain wished to take a closer look at the needs, goals, and requirements of healthcare information professionals performing complex searches of medical literature databases. Using a survey, they asked members of professional healthcare associations questions such as "how do you formulate search strategies?" and "what functionality do you value in a literature search system?". The researchers concluded that "current literature search systems offer only limited support for their requirements" and that "there is a need for improved functionality regarding the management of search strategies and the ability to search across multiple databases."
Posted on November 2, 2017
By Shawn Douglas
Journal articles
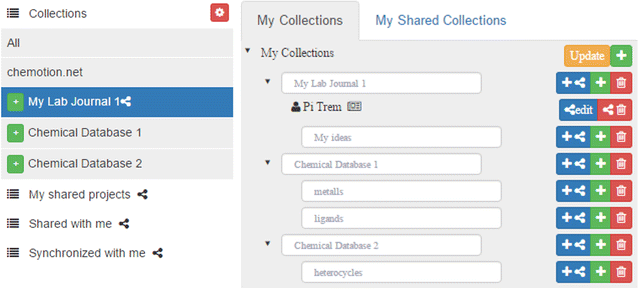
Unhappy with the electronic laboratory notebook (ELN) offerings for organic and inorganic chemists, a group of German researchers decided to take development of such an ELN into their own hands. This 2017 paper by Tremouilhac
et al. details the process of developing and implementing their own open-source solution, Chemotion ELN. After over a half year of use, the researchers state the ELN new serves "as a common infrastructure for chemistry research and [enables] chemistry researchers to build their own databases of digital information as a prerequisite for the detailed, systematic investigation and evaluation of chemical reactions and mechanisms." The group is hoping to add more functionality in time, including document generation and management, additional database query support, and an API for chemistry repositories.
Posted on October 24, 2017
By Shawn Douglas
Journal articles
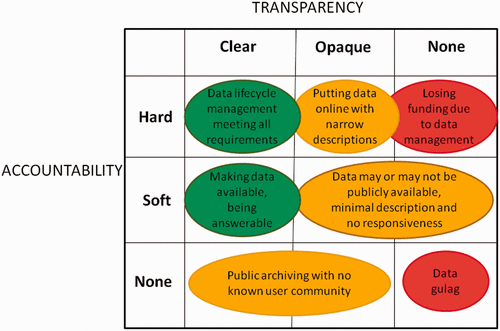
In this brief commentary article, University Corporation for Atmospheric Research's Matthew Mayernik discusses the concepts of "accountability" and "transparency" in regards to open data, conceptualizing open data "as the result of ongoing achievements, not one-time acts." In his conclusion, Mayernik notes that "good data management can happen even without sanctions," and that provision of access alone doesn't make something transparent, meaning that licenses and other complications make transparency difficult and require researchers "to account for changing expectations and requirements" of open data.
Posted on October 18, 2017
By Shawn Douglas
Journal articles
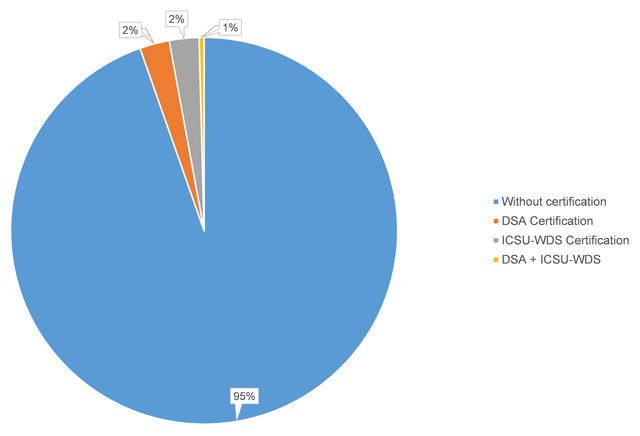
The use of publicly available data repositories is increasingly encouraged by organizations handling academic researchers' publications. Funding agencies, data organizations, and academic publishers alike are providing their researchers with lists of recommended repositories, though many aren't certified (as little as six percent of recommended are certified). Husen
et al. show that the landscape of recommended and certified data repositories is varied, and they conclude that "common ground for the standardization of data repository requirements" should be a common goal.
Posted on October 9, 2017
By Shawn Douglas
Journal articles
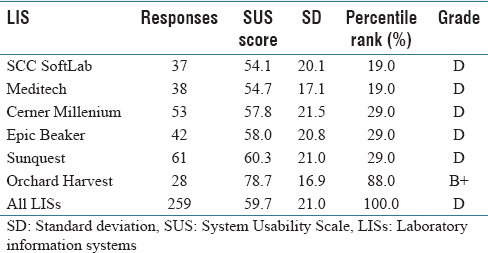
In this 2017 paper by Mathews and Marc, a picture is painted of laboratory information systems (LIS) usability, based on a structured survey with more than 250 qualifying responses: LIS design isn't particularly user-friendly. From
ad hoc laboratory reporting to quality control reporting, users consistently found several key LIS tasks either difficult or very difficult, with Orchard Harvest as the demonstrable exception. They conclude "that usability of an LIS is quite poor compared to various systems that have been evaluated using the [System Usability Scale]" though "[f]urther research is warranted to determine the root cause(s) of the difference in perceived usability" between Orchard Harvest and other discussed LIS.
 Ko et al.of the Korean BioInformation Center discuss their Closha web service for large-scale genomic data analysis in this 2018 paper published in BMC Bioinformatics. Noting a lack of rapid, cost-effective genomics workflow capable of running all the major genomic data analysis application in one pipeline, the researchers developed a hybrid system that can combine workflows. Additionally, they developed an add-on tool that handles the sheer size of genomic files and the speed with which they transfer, reaching transfer speeds "of up to 10 times that of normal FTP and HTTP." They conclude that "Closha allows genomic researchers without informatics or programming expertise to perform complex large-scale analysis with only a web browser."
Ko et al.of the Korean BioInformation Center discuss their Closha web service for large-scale genomic data analysis in this 2018 paper published in BMC Bioinformatics. Noting a lack of rapid, cost-effective genomics workflow capable of running all the major genomic data analysis application in one pipeline, the researchers developed a hybrid system that can combine workflows. Additionally, they developed an add-on tool that handles the sheer size of genomic files and the speed with which they transfer, reaching transfer speeds "of up to 10 times that of normal FTP and HTTP." They conclude that "Closha allows genomic researchers without informatics or programming expertise to perform complex large-scale analysis with only a web browser."
 In this 2018 paper published in Scientific Programming, Zhu et al. review the state of big data in the geological sciences and provide context to the challenges associated with managing that data in the cloud using China's various databases and tools as examples. Using the term "cloud-enabled geological information services" or CEGIS, they also outline the existing and new technologies that will bring together and shape how geologic data is accessed and used in the cloud. They conclude that "[w]ith the continuous development of big data technologies in addressing those challenges related to geological big data, such as the difficulties of describing and modeling geological big data with some complex characteristics, CEGIS will move towards a more mature and more intelligent direction in the future."
In this 2018 paper published in Scientific Programming, Zhu et al. review the state of big data in the geological sciences and provide context to the challenges associated with managing that data in the cloud using China's various databases and tools as examples. Using the term "cloud-enabled geological information services" or CEGIS, they also outline the existing and new technologies that will bring together and shape how geologic data is accessed and used in the cloud. They conclude that "[w]ith the continuous development of big data technologies in addressing those challenges related to geological big data, such as the difficulties of describing and modeling geological big data with some complex characteristics, CEGIS will move towards a more mature and more intelligent direction in the future."
 Reporting isn't as simple as casually placing key figures on a page; significant work should go into designing a report template, particularly those reporting specialized data , like that found in the world of pathogen genomics. Crisan et al. of the University of British Columbia and the BC Centre for Disease Control looked for evidence-based guidelines on creating reports for such a specialty — specifically for tuberculosis genomic testing — and couldn't find any. So they researched and created their own. This 2018 paper details their journey towards a final report design, concluding "that the application of human-centered design methodologies allowed us to improve not only the visual aesthetics of the final report, but also its functionality, by carefully coupling stakeholder tasks, data, and constraints to techniques from information and graphic design."
Reporting isn't as simple as casually placing key figures on a page; significant work should go into designing a report template, particularly those reporting specialized data , like that found in the world of pathogen genomics. Crisan et al. of the University of British Columbia and the BC Centre for Disease Control looked for evidence-based guidelines on creating reports for such a specialty — specifically for tuberculosis genomic testing — and couldn't find any. So they researched and created their own. This 2018 paper details their journey towards a final report design, concluding "that the application of human-centered design methodologies allowed us to improve not only the visual aesthetics of the final report, but also its functionality, by carefully coupling stakeholder tasks, data, and constraints to techniques from information and graphic design."
 This 2017 paper by Saa et al. examines the existing research literature concerning the status of moving an enterprise resource planning (ERP) system to the cloud. Noting both the many benefits of cloud for ERP and the data security drawbacks, the researchers discover the use of hybrid cloud-based ERPs by some organizations as a way to strike a balance, enabling them to "benefit from the agility and scalability of cloud-based ERP solutions while still keeping the security advantages from on-premise solutions for their mission-critical data."
This 2017 paper by Saa et al. examines the existing research literature concerning the status of moving an enterprise resource planning (ERP) system to the cloud. Noting both the many benefits of cloud for ERP and the data security drawbacks, the researchers discover the use of hybrid cloud-based ERPs by some organizations as a way to strike a balance, enabling them to "benefit from the agility and scalability of cloud-based ERP solutions while still keeping the security advantages from on-premise solutions for their mission-critical data."
 A major goal among researchers in the age of big data is to improve data sharing and collaborative efforts. Kindler et al. note, for example, that "specialized techniques for the production and analysis of viral metagenomes remains in a subset of labs," and other researchers likely don't know about them. As such, the authors of this 2017 paper in F1000Research discuss the enhancement of the protocols.io online collaborative space to include more social elements "for scientists to share improvements and corrections to protocols so that others are not continuously re-discovering knowledge that scientists have not had the time or wear-with-all to publish." With its many connection points, they conclude, the software update "will allow the forum to evolve naturally given rapidly developing trends and new protocols."
A major goal among researchers in the age of big data is to improve data sharing and collaborative efforts. Kindler et al. note, for example, that "specialized techniques for the production and analysis of viral metagenomes remains in a subset of labs," and other researchers likely don't know about them. As such, the authors of this 2017 paper in F1000Research discuss the enhancement of the protocols.io online collaborative space to include more social elements "for scientists to share improvements and corrections to protocols so that others are not continuously re-discovering knowledge that scientists have not had the time or wear-with-all to publish." With its many connection points, they conclude, the software update "will allow the forum to evolve naturally given rapidly developing trends and new protocols."
 This brief paper by Ganzinger and Knaup examines the state of systems medicine, where multiple medical data streams are merged, analyzed, modeled, etc. to further how we diagnose and treat disease. They discuss the dynamic nature of disease knowledge and clinical data, as well as the problems that arise from integrating omics data into systems medicine, under the umbrella of integrating the knowledge into a usable format in tools like a clinical decision support system. They conclude that though with many benefits, "special care has to be taken to address inherent dynamics of data that are used for systems medicine; over time the number of available health records will increase and treatment approaches will change." They add that their data management model is flexible and can be used with other data management tools.
This brief paper by Ganzinger and Knaup examines the state of systems medicine, where multiple medical data streams are merged, analyzed, modeled, etc. to further how we diagnose and treat disease. They discuss the dynamic nature of disease knowledge and clinical data, as well as the problems that arise from integrating omics data into systems medicine, under the umbrella of integrating the knowledge into a usable format in tools like a clinical decision support system. They conclude that though with many benefits, "special care has to be taken to address inherent dynamics of data that are used for systems medicine; over time the number of available health records will increase and treatment approaches will change." They add that their data management model is flexible and can be used with other data management tools.
 Published in early 2017, this paper by Albert et al. discusses the development process of an accreditation program — the Strengthening Tuberculosis Laboratory Management Toward Accreditation or TB SLMTA — dedicated to better implementation of quality management systems (QMS) in tuberculosis laboratories around the world. The authors discuss the development of the curriculum, accreditation tools, and roll-out across 10 countries and 37 laboratories. They conclude that the training and mentoring program is increasingly vital for tuberculosis labs, "building a foundation toward further quality improvement toward achieving accreditation ... on the African continent and beyond."
Published in early 2017, this paper by Albert et al. discusses the development process of an accreditation program — the Strengthening Tuberculosis Laboratory Management Toward Accreditation or TB SLMTA — dedicated to better implementation of quality management systems (QMS) in tuberculosis laboratories around the world. The authors discuss the development of the curriculum, accreditation tools, and roll-out across 10 countries and 37 laboratories. They conclude that the training and mentoring program is increasingly vital for tuberculosis labs, "building a foundation toward further quality improvement toward achieving accreditation ... on the African continent and beyond."
 Dereplication is a chemical screening process of separation and purification that eliminates known and studied constituents, leaving other novel metabolites for future study. This is an important part of pharmaceutical and natural product development, requiring appropriate data management and retrieval for researchers around the world. However, Scotti et al. found a lack of secondary metabolite databases that met their needs. Noting a need for complex searches, structure management, visualizations, and taxonomic rank in one package, they developed SistematX, "a modern and innovative web interface." They conclude the end result is a system that "provides a wealth of useful information for the scientific community about natural products, highlighting the location of species from which the compounds were isolated."
Dereplication is a chemical screening process of separation and purification that eliminates known and studied constituents, leaving other novel metabolites for future study. This is an important part of pharmaceutical and natural product development, requiring appropriate data management and retrieval for researchers around the world. However, Scotti et al. found a lack of secondary metabolite databases that met their needs. Noting a need for complex searches, structure management, visualizations, and taxonomic rank in one package, they developed SistematX, "a modern and innovative web interface." They conclude the end result is a system that "provides a wealth of useful information for the scientific community about natural products, highlighting the location of species from which the compounds were isolated."
 Metadata capture is important to scientific reproducibility argue Zehl et al., particularly in regards to "the experiment, the data, and the applied preprocessing steps on the data." In this 2016 paper, however, the researchers demonstrate that metadata capture is not necessarily a simple task for an experimental laboratory; diverse datasets, specialized workflows, and lack of knowledge of supporting software tools all make the challenge of metadata capture more difficult. After demonstrating use cases representative of their neurophysiology lab and make a case for the open metadata Markup Language (odML), the researchers conclude that "[r]eadily available tools to support metadata management, such as odML, are a vital component in constructing [data and metadata management] workflows" in laboratories of all types.
Metadata capture is important to scientific reproducibility argue Zehl et al., particularly in regards to "the experiment, the data, and the applied preprocessing steps on the data." In this 2016 paper, however, the researchers demonstrate that metadata capture is not necessarily a simple task for an experimental laboratory; diverse datasets, specialized workflows, and lack of knowledge of supporting software tools all make the challenge of metadata capture more difficult. After demonstrating use cases representative of their neurophysiology lab and make a case for the open metadata Markup Language (odML), the researchers conclude that "[r]eadily available tools to support metadata management, such as odML, are a vital component in constructing [data and metadata management] workflows" in laboratories of all types.
 In this 2017 paper by Curtin University's Cameron Neylon, the concepts of research data management (RDM) and research data sharing (RDS) are solidified and identified as increasingly common practices for funded research enterprises, particularly public-facing ones. But what of the implementation of these practices and the challenges associated with them? Neylon finds more than expected in his study, identifying sharp contrast among "those who saw [data management] requirements as a necessary part of raising issues among researchers and those concerned that this was leading to a compliance culture, where data management was seen as a required administrative exercise rather than an integral part of research practice." Neylon concludes with three key recommendations for researchers in general should follow to further shape how RDM and RDS are approached in research communities.
In this 2017 paper by Curtin University's Cameron Neylon, the concepts of research data management (RDM) and research data sharing (RDS) are solidified and identified as increasingly common practices for funded research enterprises, particularly public-facing ones. But what of the implementation of these practices and the challenges associated with them? Neylon finds more than expected in his study, identifying sharp contrast among "those who saw [data management] requirements as a necessary part of raising issues among researchers and those concerned that this was leading to a compliance culture, where data management was seen as a required administrative exercise rather than an integral part of research practice." Neylon concludes with three key recommendations for researchers in general should follow to further shape how RDM and RDS are approached in research communities.
 In this technical note from the University of Iowa Hospitals and Clinics, Imborek et al. walk through the benefits and challenges of evaluating, implementing, and assessing preferred name and gender identity into its various laboratory systems and workflows. As preferred name has been an important point "in providing inclusion toward a class of patients that have historically been disenfranchised from the healthcare system" — primarily transgender patients — awareness is increasing at many levels about not only the inclusion of preferred name but also the complications that arise from it. From customizing software to billing and coding issues, from blood donor issues to laboratory test challenges, the group concludes that despite the "major undertaking [that] required the combined effort of many people from different teams," the efforts provide new opportunities for the field of pathology.
In this technical note from the University of Iowa Hospitals and Clinics, Imborek et al. walk through the benefits and challenges of evaluating, implementing, and assessing preferred name and gender identity into its various laboratory systems and workflows. As preferred name has been an important point "in providing inclusion toward a class of patients that have historically been disenfranchised from the healthcare system" — primarily transgender patients — awareness is increasing at many levels about not only the inclusion of preferred name but also the complications that arise from it. From customizing software to billing and coding issues, from blood donor issues to laboratory test challenges, the group concludes that despite the "major undertaking [that] required the combined effort of many people from different teams," the efforts provide new opportunities for the field of pathology.
 Sure, there's plenty of talk about big data in the research and clinical realms, but what of smarter data, gained from using business process management (BPM) tools? That's the direction Andellini et al. went in after looking at the kidney transplantation cases at Bambino Gesù Children’s Hospital, realizing the potential for improving clinical decisions, scheduling of patient appointments, and reducing human errors in workflow. The group concluded "that the usage of BPM in the management of kidney transplantation leads to real benefits in terms of resource optimization and quality improvement," and that a "combination of a BPM-based platform with a service-oriented architecture could represent a revolution in clinical pathway management."
Sure, there's plenty of talk about big data in the research and clinical realms, but what of smarter data, gained from using business process management (BPM) tools? That's the direction Andellini et al. went in after looking at the kidney transplantation cases at Bambino Gesù Children’s Hospital, realizing the potential for improving clinical decisions, scheduling of patient appointments, and reducing human errors in workflow. The group concluded "that the usage of BPM in the management of kidney transplantation leads to real benefits in terms of resource optimization and quality improvement," and that a "combination of a BPM-based platform with a service-oriented architecture could represent a revolution in clinical pathway management."
 In this 2017 article published in JMIR Medical Informatics, Russell-Rose and Chamberlain wished to take a closer look at the needs, goals, and requirements of healthcare information professionals performing complex searches of medical literature databases. Using a survey, they asked members of professional healthcare associations questions such as "how do you formulate search strategies?" and "what functionality do you value in a literature search system?". The researchers concluded that "current literature search systems offer only limited support for their requirements" and that "there is a need for improved functionality regarding the management of search strategies and the ability to search across multiple databases."
In this 2017 article published in JMIR Medical Informatics, Russell-Rose and Chamberlain wished to take a closer look at the needs, goals, and requirements of healthcare information professionals performing complex searches of medical literature databases. Using a survey, they asked members of professional healthcare associations questions such as "how do you formulate search strategies?" and "what functionality do you value in a literature search system?". The researchers concluded that "current literature search systems offer only limited support for their requirements" and that "there is a need for improved functionality regarding the management of search strategies and the ability to search across multiple databases."
 Unhappy with the electronic laboratory notebook (ELN) offerings for organic and inorganic chemists, a group of German researchers decided to take development of such an ELN into their own hands. This 2017 paper by Tremouilhac et al. details the process of developing and implementing their own open-source solution, Chemotion ELN. After over a half year of use, the researchers state the ELN new serves "as a common infrastructure for chemistry research and [enables] chemistry researchers to build their own databases of digital information as a prerequisite for the detailed, systematic investigation and evaluation of chemical reactions and mechanisms." The group is hoping to add more functionality in time, including document generation and management, additional database query support, and an API for chemistry repositories.
Unhappy with the electronic laboratory notebook (ELN) offerings for organic and inorganic chemists, a group of German researchers decided to take development of such an ELN into their own hands. This 2017 paper by Tremouilhac et al. details the process of developing and implementing their own open-source solution, Chemotion ELN. After over a half year of use, the researchers state the ELN new serves "as a common infrastructure for chemistry research and [enables] chemistry researchers to build their own databases of digital information as a prerequisite for the detailed, systematic investigation and evaluation of chemical reactions and mechanisms." The group is hoping to add more functionality in time, including document generation and management, additional database query support, and an API for chemistry repositories.
 In this brief commentary article, University Corporation for Atmospheric Research's Matthew Mayernik discusses the concepts of "accountability" and "transparency" in regards to open data, conceptualizing open data "as the result of ongoing achievements, not one-time acts." In his conclusion, Mayernik notes that "good data management can happen even without sanctions," and that provision of access alone doesn't make something transparent, meaning that licenses and other complications make transparency difficult and require researchers "to account for changing expectations and requirements" of open data.
In this brief commentary article, University Corporation for Atmospheric Research's Matthew Mayernik discusses the concepts of "accountability" and "transparency" in regards to open data, conceptualizing open data "as the result of ongoing achievements, not one-time acts." In his conclusion, Mayernik notes that "good data management can happen even without sanctions," and that provision of access alone doesn't make something transparent, meaning that licenses and other complications make transparency difficult and require researchers "to account for changing expectations and requirements" of open data.
 The use of publicly available data repositories is increasingly encouraged by organizations handling academic researchers' publications. Funding agencies, data organizations, and academic publishers alike are providing their researchers with lists of recommended repositories, though many aren't certified (as little as six percent of recommended are certified). Husen et al. show that the landscape of recommended and certified data repositories is varied, and they conclude that "common ground for the standardization of data repository requirements" should be a common goal.
The use of publicly available data repositories is increasingly encouraged by organizations handling academic researchers' publications. Funding agencies, data organizations, and academic publishers alike are providing their researchers with lists of recommended repositories, though many aren't certified (as little as six percent of recommended are certified). Husen et al. show that the landscape of recommended and certified data repositories is varied, and they conclude that "common ground for the standardization of data repository requirements" should be a common goal.
 In this 2017 paper by Mathews and Marc, a picture is painted of laboratory information systems (LIS) usability, based on a structured survey with more than 250 qualifying responses: LIS design isn't particularly user-friendly. From ad hoc laboratory reporting to quality control reporting, users consistently found several key LIS tasks either difficult or very difficult, with Orchard Harvest as the demonstrable exception. They conclude "that usability of an LIS is quite poor compared to various systems that have been evaluated using the [System Usability Scale]" though "[f]urther research is warranted to determine the root cause(s) of the difference in perceived usability" between Orchard Harvest and other discussed LIS.
In this 2017 paper by Mathews and Marc, a picture is painted of laboratory information systems (LIS) usability, based on a structured survey with more than 250 qualifying responses: LIS design isn't particularly user-friendly. From ad hoc laboratory reporting to quality control reporting, users consistently found several key LIS tasks either difficult or very difficult, with Orchard Harvest as the demonstrable exception. They conclude "that usability of an LIS is quite poor compared to various systems that have been evaluated using the [System Usability Scale]" though "[f]urther research is warranted to determine the root cause(s) of the difference in perceived usability" between Orchard Harvest and other discussed LIS.
















