Posted on July 10, 2017
By Shawn Douglas
Journal articles
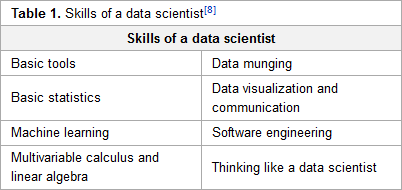
Like many other fields of science, earth science is increasingly swimming in data. Unlike other fields, the discussion of earth science data and its analysis hasn't been as vigorous, comparatively speaking. Kempler and Mathews of NASA speak of their efforts within the Earth Science Information Partners' (ESIP) earth science data analytics (ESDA) program in this 2017 paper published in
Data Science Journal. The duo shares their experiences and provides a set of techniques and skills typical to ESDA "that is useful in articulating data management and research needs, as well as a working list of techniques and skills relevant to the different types of ESDA."
Posted on July 5, 2017
By John Jones
Journal articles
This brief paper published in
BMC Bioinformatics provides a sociological examination of the world of bioinformatics and how it's perceived institutionally. Bartlett
et al. argue that institutionally while less focus is place on bioinformatics processes and more on the data input and output, putting the contributions of bioinformaticists into a "black box," losing scientific credit in the process. The researchers conclude that "[i]n the pursuit of relevance and impact, future scientific careers will increasingly involve playing the role of a fractional scientist ... combining a variety of expertise and epistemic aspirations..." to become "tomorrow's bioinformatic scientists."
Posted on June 27, 2017
By John Jones
Journal articles
In this 2017 journal article published in the
Data Science Journal, data scientist Sabina Leonelli reflects on a paradigm shift in biology where "specific data production technologies [are used] as proxy for assessing data quality," creating problems along the way, particularly for the open data movement. Leonelli's major concern: "Ethnographic research carried out in such environments evidences a widespread fear among researchers that providing extensive information about their experimental set-up will affect the perceived quality of their data, making their findings vulnerable to criticisms by better-resourced peers," hindering data and provenance sharing. And the conclusion? Endorsing specific data production and management technologies as indicators of data quality can cloud the goals of open data initiatives.
Posted on June 20, 2017
By John Jones
Journal articles
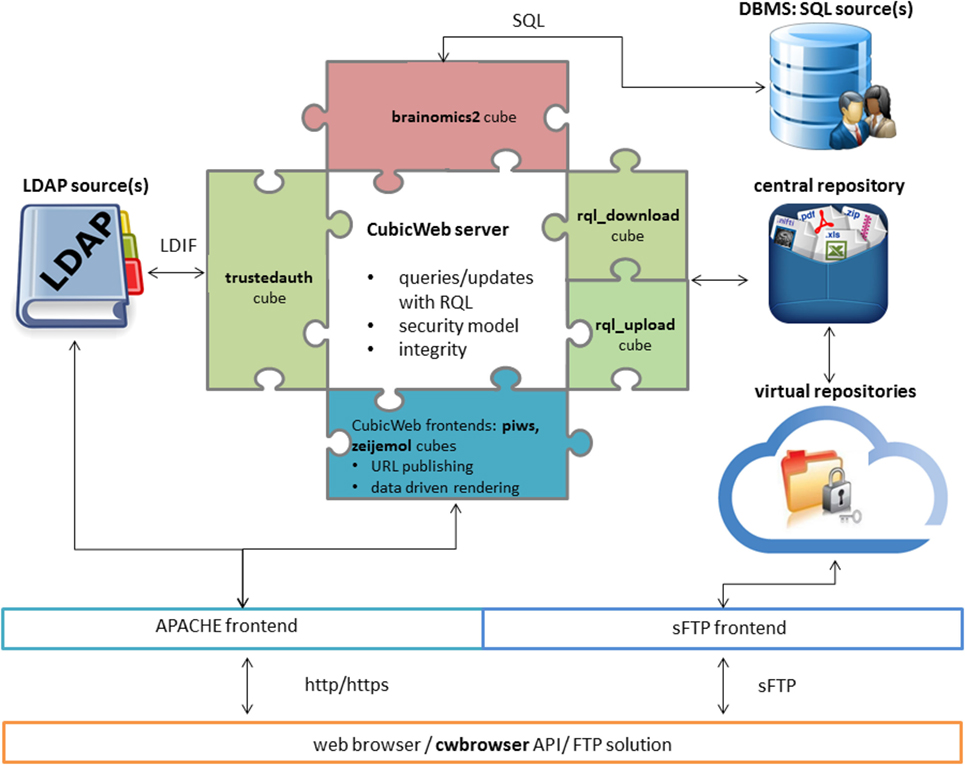
In this 2017 article published in
Frontiers in Neuroinformatics, Grigis
et al., like many before them, note that data in scientific areas of research such as genetics, imaging, and the social sciences has become "massive, heterogeneous, and complex." Their solution is a Python-based one that integrates the CubicWeb open-source semantic framework and other tools "to overcome the challenges associated with data sharing and collaborative requirements" found in population imaging studies. The resulting data sharing service (DSS) proves to be flexible, integratable, and expandable based on demand, they conclude.
Posted on June 13, 2017
By John Jones
Journal articles
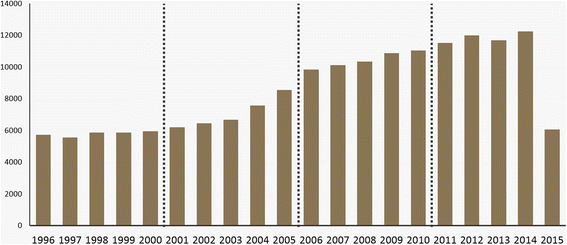
Wikipedia defines bibliometrics as a "statistical analysis of written publications, such as books or articles." Related to information and library science, bibliometrics has been helping researchers make better sense of the trends and impacts made across numerous fields. In this 2017 paper, Heo
et al. use bibliometric methods new and old to examine the field of bioinformatics via related journals over a period of 20 years to better understand how the field has changed in that time. They conclude that "the characteristics of the bioinformatics field become more distinct and more specific, and the supporting role of peripheral fields of bioinformatics, such as conceptual, mathematical, and systems biology, gradually increases over time, though the core fields of proteomics, genomics, and genetics are still the major topics."
Posted on June 6, 2017
By John Jones
Journal articles
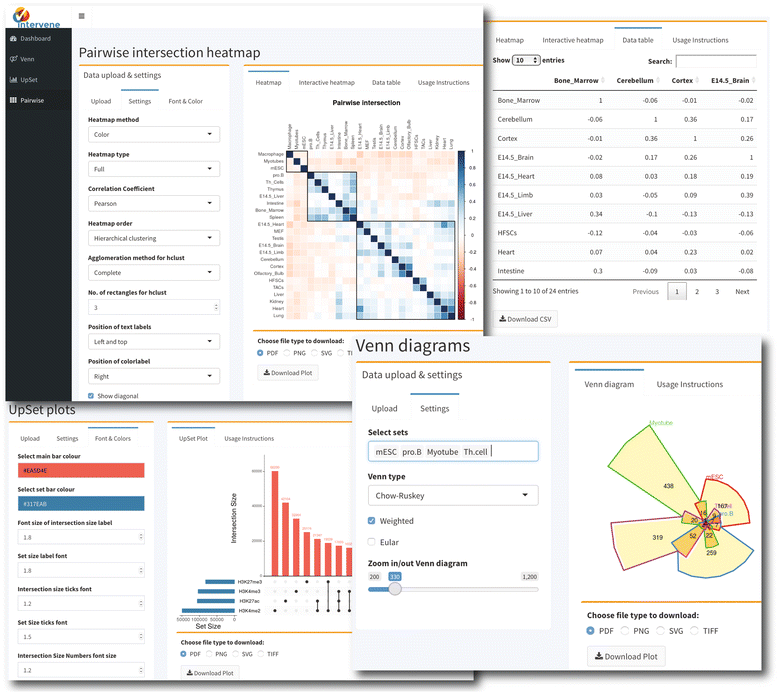
As Khan and Mathelier note in their abstract, one of the more common tasks of a bioinformatician is to take lists of genomes or genomic regions from high-throughput sequencing and compare them visually. Noting the lack of a comprehensive tool to visualize such complex datasets, the authors developed Intervene, a tool for computing intersections of multiple genomic and list sets. They conclude that "Intervene is the first tool to provide three types of visualization approaches for multiple sets of gene or genomic intervals," and they have made the the source code, web app, and documentation freely available to the public.
Posted on May 31, 2017
By John Jones
Journal articles
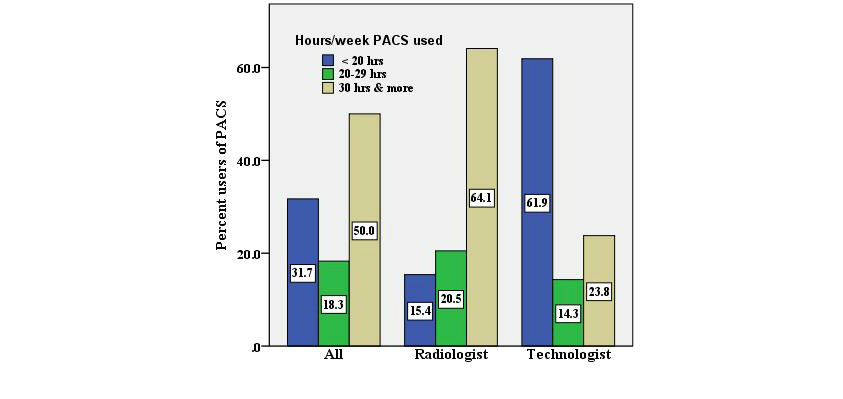
The picture archiving and communication system (PACS) is an increasingly important information management component of hospitals and medical centers, allowing for the digital acquisition, archiving, communication, retrieval, processing, distribution, and display of medical images. But do staff members using it find that a PACS makes their job easier and more effective? This journal article by Buabbas
et al. represents another attempt by medical researchers to quantify and qualify the impact of the PACS on radiologists and technologists using the system. In their case, the authors concluded that "[d]espite some of the technical limitations of the infrastructure, most of the respondents rated the system positively and as user-friendly" but, like any information system, there are still a few areas of improvement that need attention.
Posted on May 23, 2017
By John Jones
Journal articles
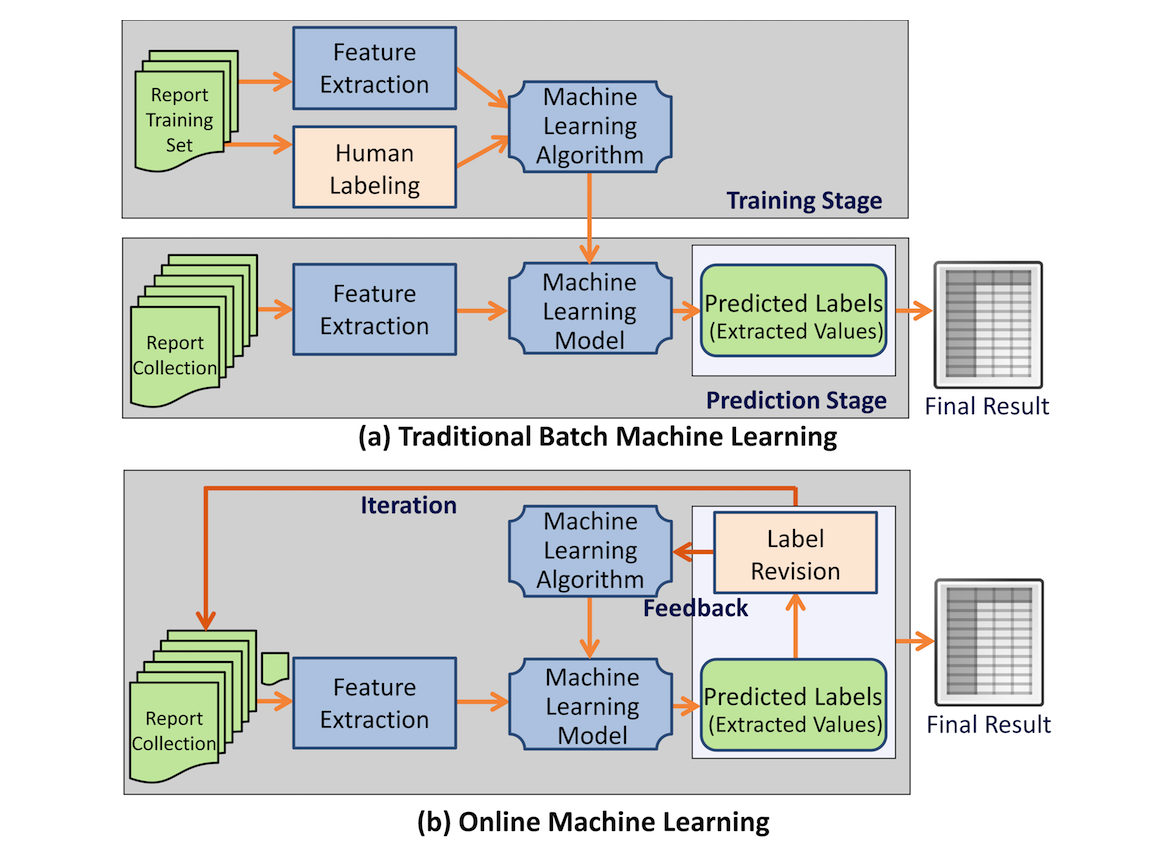
Even in the digital realm (think electronic medical records), extracting usable information from narrated medical reports can be a challenge given heterogeneous data structures and vocabularies. While many systems have been created over the years to tackle this task, researchers from Emory and Stony Brook University have taken a different approach: online learning. Here Zheng
et al. present their methodology and findings associated with their Information and Data Extraction using Adaptive Online Learning (IDEAL-X) system, concluding that "the online learning–based method combined with controlled vocabularies for data extraction from reports with various structural patterns ... is highly effective."
Posted on May 15, 2017
By John Jones
Journal articles
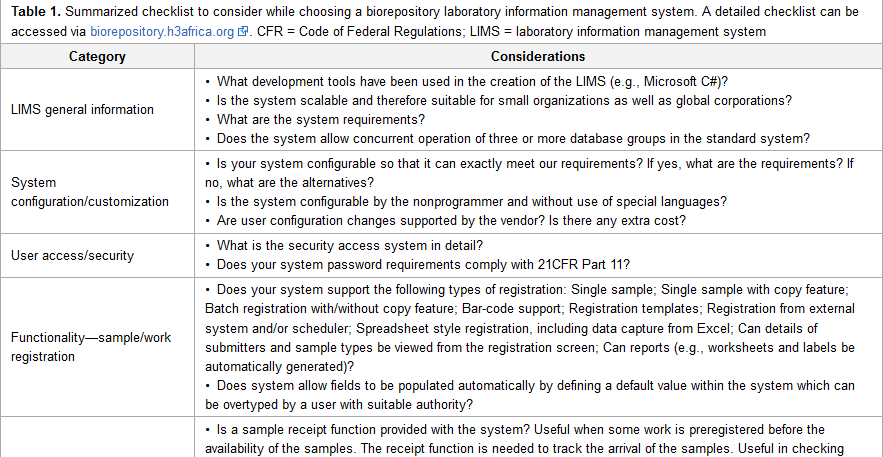
What's important for a biorepository laboratory information management system (LIMS), and what options are out there? What unique constraints in Africa make that selection more difficult? This brief 2017 paper from the Human Heredity and Health in Africa (H3Africa) Consortium outlines their take on finding the right LIMS solution for three of their regional biorepositories in Africa. The group emphasizes in the end that "[c]hoosing a LIMS in low- and middle-income countries requires careful consideration of the various factors that could affect its successful and sustainable deployment and use."
Posted on May 10, 2017
By John Jones
Journal articles

This journal article, published in
Biopreservation and Biobanking in early 2017, presents the development philosophy and implementation of a custom-modified version of Bika LIMS called Baobab LIMS, designed for biobank clients and researchers. Bendou
et al., who enlisted customization help directly from Bika Lab System, describe how "[t]he need to implement biobank standard operation procedures as well as stimulate the use of standards for biobank data representation motivated the implementation of Baobab LIMS, an open-source LIMS for biobanking." The group concludes that while the open-source LIMS is quite usable as is, it will require further development of more "generic and configurable workflows." Despite this, the authors anticipate the software to be useful to the biobanking community.
Posted on May 2, 2017
By John Jones
Journal articles
Most scientists know that much of the data created in academic research efforts ends up being locked away in silos, difficult to share with others. But what are scientists doing about? In this 2016 paper published in
Scientific Data, Wilkinson
et al. outline a distinct set of principles created towards reducing the silos of information: the FAIR Principles. The authors state the primary goal of the FAIR Principles is to "put specific emphasis on enhancing the ability of machines to automatically find and use the data, in addition to supporting its reuse by individuals." After describing the principles and giving examples of projects that adhere to them, the authors conclude that the principles have the potential to "guide the implementation of the most basic levels of good Data Management and Stewardship practice, thus helping researchers adhere to the expectations and requirements of their funding agencies."
Posted on April 26, 2017
By John Jones
Journal articles

The problem? Disparate data sources, from weather and wave forecasts to navigation charts and natural hazard assessments, made oceanography research in Southern Italy more cumbersome. Solution? Create a secure, standardized, and interoperable data platform that can merge all that and other information together into one powerful and easy-to-use platform. Thus the TESSA (Development of Technology for Situational Sea Awareness) program was born. D'Anca
et al. discuss the creation and use of TESSA as a geospatial tool that merges real-time and archived data to help researchers in Southern Italy. The authors conclude that TESSA is "a valid prototype easily adopted to provide an efficient dissemination of maritime data and a consolidation of the management of operational oceanographic activities," even in other parts of the world.
Posted on April 17, 2017
By John Jones
Journal articles

In development since at least the summer of 2009, the open-source MASTR-MS laboratory information management system (LIMS) was designed to better handle the data and metadata of metabolomics, the study of an entity's metabolites. In this 2017 paper published in
Metabolomics, the development team of MASTR-MS discuss the current state of their LIMS, how it's being used, and what the future holds for it. They conclude by stating the software's "comprehensive functions and features enable researchers and facilities to effectively manage a wide range of different project and experimental data types, and it facilitate the mining of new and existing [metabolomic] datasets."
Posted on April 10, 2017
By John Jones
Journal articles

When designing something as simple as a menu of laboratory tests into a piece of clinical software, it's relatively easy to not think of the ramifications of the contents of such a menu. In this 2017 article published in
BMC Medical Informatics and Decision Making, Martins
et al. argue that there are consequences to what's included in a laboratory test drop-down menu, primarily that the presence — or lack thereof — of a test type may influence how frequently that test is prescribed. The group concludes that "[r]emoving unnecessary tests from a quick shortcut menu of the diagnosis and laboratory tests ordering system had a significant impact and reduced unnecessary prescription of tests," which in turn led "to the reduction of negative patient effects and to the reduction of unnecessary costs."
Posted on April 5, 2017
By John Jones
Journal articles
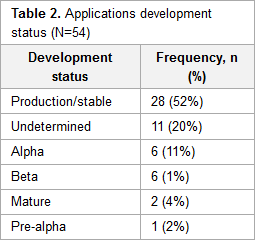
In this journal article published in
JMIR Medical Informatics in 2017, Alsaffar
et al. review research from mid-2014 that looked at the state of open-source electronic health record (EHR) systems, primarily via SourceForge. The authors, noting a lack of research concerning the demographics and motivation of open-source EHR projects, present their finding, concluding that "lack of a corporate entity in most F/OSS EHR projects translates to a marginal capacity to market the respective F/OSS system and to navigate [HITECH] certification."
Posted on March 27, 2017
By John Jones
Journal articles

In this 2017 paper published in
BMC Bioinformatics, Eyal-Altman
et al. explain the use and benefits of their KNIME-based cancer outcome analysis software PCM-SABRE (Precision Cancer Medicine - Survival Analysis Benchmarking, Reporting and Evaluation). The group demonstrates its effectiveness by reconstructing the previous work of Chou
et al. and showing how the results necessitate the tool for better reproducibility. The researchers conclude that when used effectively, PCM-Sabre's "resulting pipeline can be shared with others in an intuitive yet executable way, which will improve, if adopted by other investigators, the comparability and interpretability of future works attempting to predict patient survival from gene expression data."
Posted on March 22, 2017
By John Jones
Journal articles
This journal article in
PLOS Computational Biology's long-running
Ten Simple Rules series goes back to 2013, when a collaborative group of eight authors from around the globe pooled their thoughts together on the topic of open science and collaborative R&D. The conversations (linked to in this article) provide context and insight into the various projects — from the Gene Wiki initiative to the Open Source Drug Discovery (OSDD) project — that have required significant deviation of thought from the traditional company view of conducting business. From "lead as a coach, not a CEO" to "grow the commons," the article's authors provide their thoughts on what best makes for collaborative and open science projects.
Posted on March 14, 2017
By John Jones
Journal articles
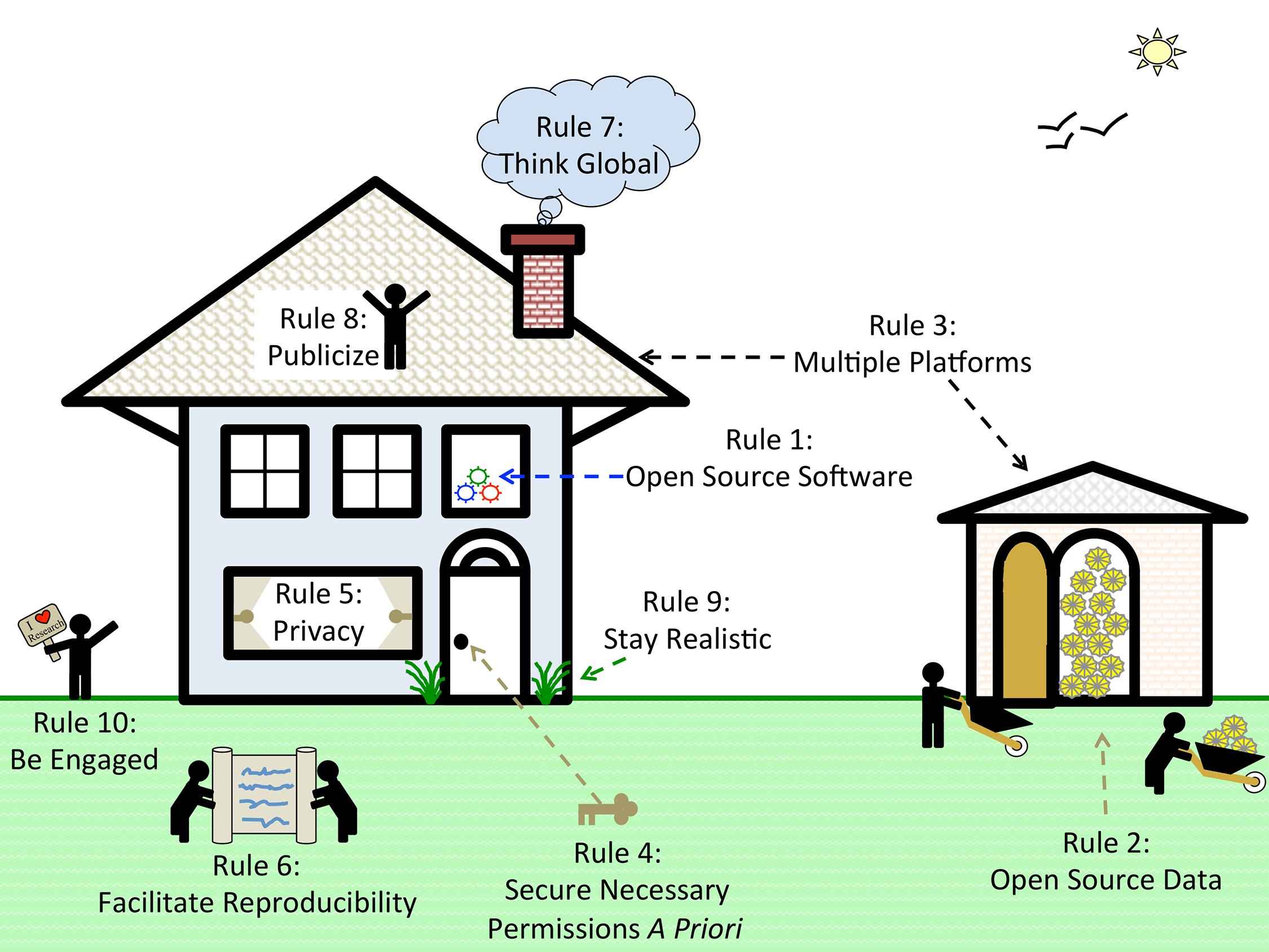
In yet another installment of
PLOS Computational Biology's
Ten Simple Rules series, Boland
et al. of Columbia University and the Broad Institute of MIT and Harvard share their thoughts and experiences with multi-site collaborations and data sharing. The group provides practical tips for making data sharing easier and more successful, strengthening collaborations and the scientific process.
Posted on March 7, 2017
By John Jones
Journal articles
This is another entry in
PLOS Computational Biology's long-running
Ten Simple Rules series, which attempts to break down computational biology / bioinformatics topics (that relate to the informatics side) down into a digestible and cited format. This 2017 entry by List
et al. looks at the typical problems associated with computational biology software development and attempts to provide a clear approach for more usable, efficient software. The authors conclude that despite following these 10 rules, there's more to be done: "...effort is required from both users and developers to further improve a tool. Even engaging with only a few users ... is likely to have a large impact on usability."
Posted on March 1, 2017
By John Jones
Journal articles
In this brief paper by Rumbold and Pierscionek, the implications and theoretical impact of the European Union's General Data Protection Regulation are discussed. Addressing in particular claims that the new "consent requirements ... would severely restrict medical data research," the researchers break down the law that goes into effect in 2018, including anonymization issues, consent issues, and data sharing issues that will potentially affect biomedical data research. They conclude the impact will by minimal: "The GDPR will facilitate medical research, except where it is research not considered in the public interest. In that case, more demanding requirements for anonymization will entail either true anonymization or consent."
 Like many other fields of science, earth science is increasingly swimming in data. Unlike other fields, the discussion of earth science data and its analysis hasn't been as vigorous, comparatively speaking. Kempler and Mathews of NASA speak of their efforts within the Earth Science Information Partners' (ESIP) earth science data analytics (ESDA) program in this 2017 paper published in Data Science Journal. The duo shares their experiences and provides a set of techniques and skills typical to ESDA "that is useful in articulating data management and research needs, as well as a working list of techniques and skills relevant to the different types of ESDA."
Like many other fields of science, earth science is increasingly swimming in data. Unlike other fields, the discussion of earth science data and its analysis hasn't been as vigorous, comparatively speaking. Kempler and Mathews of NASA speak of their efforts within the Earth Science Information Partners' (ESIP) earth science data analytics (ESDA) program in this 2017 paper published in Data Science Journal. The duo shares their experiences and provides a set of techniques and skills typical to ESDA "that is useful in articulating data management and research needs, as well as a working list of techniques and skills relevant to the different types of ESDA."
 In this 2017 article published in Frontiers in Neuroinformatics, Grigis et al., like many before them, note that data in scientific areas of research such as genetics, imaging, and the social sciences has become "massive, heterogeneous, and complex." Their solution is a Python-based one that integrates the CubicWeb open-source semantic framework and other tools "to overcome the challenges associated with data sharing and collaborative requirements" found in population imaging studies. The resulting data sharing service (DSS) proves to be flexible, integratable, and expandable based on demand, they conclude.
In this 2017 article published in Frontiers in Neuroinformatics, Grigis et al., like many before them, note that data in scientific areas of research such as genetics, imaging, and the social sciences has become "massive, heterogeneous, and complex." Their solution is a Python-based one that integrates the CubicWeb open-source semantic framework and other tools "to overcome the challenges associated with data sharing and collaborative requirements" found in population imaging studies. The resulting data sharing service (DSS) proves to be flexible, integratable, and expandable based on demand, they conclude.
 Wikipedia defines bibliometrics as a "statistical analysis of written publications, such as books or articles." Related to information and library science, bibliometrics has been helping researchers make better sense of the trends and impacts made across numerous fields. In this 2017 paper, Heo et al. use bibliometric methods new and old to examine the field of bioinformatics via related journals over a period of 20 years to better understand how the field has changed in that time. They conclude that "the characteristics of the bioinformatics field become more distinct and more specific, and the supporting role of peripheral fields of bioinformatics, such as conceptual, mathematical, and systems biology, gradually increases over time, though the core fields of proteomics, genomics, and genetics are still the major topics."
Wikipedia defines bibliometrics as a "statistical analysis of written publications, such as books or articles." Related to information and library science, bibliometrics has been helping researchers make better sense of the trends and impacts made across numerous fields. In this 2017 paper, Heo et al. use bibliometric methods new and old to examine the field of bioinformatics via related journals over a period of 20 years to better understand how the field has changed in that time. They conclude that "the characteristics of the bioinformatics field become more distinct and more specific, and the supporting role of peripheral fields of bioinformatics, such as conceptual, mathematical, and systems biology, gradually increases over time, though the core fields of proteomics, genomics, and genetics are still the major topics."
 As Khan and Mathelier note in their abstract, one of the more common tasks of a bioinformatician is to take lists of genomes or genomic regions from high-throughput sequencing and compare them visually. Noting the lack of a comprehensive tool to visualize such complex datasets, the authors developed Intervene, a tool for computing intersections of multiple genomic and list sets. They conclude that "Intervene is the first tool to provide three types of visualization approaches for multiple sets of gene or genomic intervals," and they have made the the source code, web app, and documentation freely available to the public.
As Khan and Mathelier note in their abstract, one of the more common tasks of a bioinformatician is to take lists of genomes or genomic regions from high-throughput sequencing and compare them visually. Noting the lack of a comprehensive tool to visualize such complex datasets, the authors developed Intervene, a tool for computing intersections of multiple genomic and list sets. They conclude that "Intervene is the first tool to provide three types of visualization approaches for multiple sets of gene or genomic intervals," and they have made the the source code, web app, and documentation freely available to the public.
 The picture archiving and communication system (PACS) is an increasingly important information management component of hospitals and medical centers, allowing for the digital acquisition, archiving, communication, retrieval, processing, distribution, and display of medical images. But do staff members using it find that a PACS makes their job easier and more effective? This journal article by Buabbas et al. represents another attempt by medical researchers to quantify and qualify the impact of the PACS on radiologists and technologists using the system. In their case, the authors concluded that "[d]espite some of the technical limitations of the infrastructure, most of the respondents rated the system positively and as user-friendly" but, like any information system, there are still a few areas of improvement that need attention.
The picture archiving and communication system (PACS) is an increasingly important information management component of hospitals and medical centers, allowing for the digital acquisition, archiving, communication, retrieval, processing, distribution, and display of medical images. But do staff members using it find that a PACS makes their job easier and more effective? This journal article by Buabbas et al. represents another attempt by medical researchers to quantify and qualify the impact of the PACS on radiologists and technologists using the system. In their case, the authors concluded that "[d]espite some of the technical limitations of the infrastructure, most of the respondents rated the system positively and as user-friendly" but, like any information system, there are still a few areas of improvement that need attention.
 Even in the digital realm (think electronic medical records), extracting usable information from narrated medical reports can be a challenge given heterogeneous data structures and vocabularies. While many systems have been created over the years to tackle this task, researchers from Emory and Stony Brook University have taken a different approach: online learning. Here Zheng et al. present their methodology and findings associated with their Information and Data Extraction using Adaptive Online Learning (IDEAL-X) system, concluding that "the online learning–based method combined with controlled vocabularies for data extraction from reports with various structural patterns ... is highly effective."
Even in the digital realm (think electronic medical records), extracting usable information from narrated medical reports can be a challenge given heterogeneous data structures and vocabularies. While many systems have been created over the years to tackle this task, researchers from Emory and Stony Brook University have taken a different approach: online learning. Here Zheng et al. present their methodology and findings associated with their Information and Data Extraction using Adaptive Online Learning (IDEAL-X) system, concluding that "the online learning–based method combined with controlled vocabularies for data extraction from reports with various structural patterns ... is highly effective."
 What's important for a biorepository laboratory information management system (LIMS), and what options are out there? What unique constraints in Africa make that selection more difficult? This brief 2017 paper from the Human Heredity and Health in Africa (H3Africa) Consortium outlines their take on finding the right LIMS solution for three of their regional biorepositories in Africa. The group emphasizes in the end that "[c]hoosing a LIMS in low- and middle-income countries requires careful consideration of the various factors that could affect its successful and sustainable deployment and use."
What's important for a biorepository laboratory information management system (LIMS), and what options are out there? What unique constraints in Africa make that selection more difficult? This brief 2017 paper from the Human Heredity and Health in Africa (H3Africa) Consortium outlines their take on finding the right LIMS solution for three of their regional biorepositories in Africa. The group emphasizes in the end that "[c]hoosing a LIMS in low- and middle-income countries requires careful consideration of the various factors that could affect its successful and sustainable deployment and use."
 This journal article, published in Biopreservation and Biobanking in early 2017, presents the development philosophy and implementation of a custom-modified version of Bika LIMS called Baobab LIMS, designed for biobank clients and researchers. Bendou et al., who enlisted customization help directly from Bika Lab System, describe how "[t]he need to implement biobank standard operation procedures as well as stimulate the use of standards for biobank data representation motivated the implementation of Baobab LIMS, an open-source LIMS for biobanking." The group concludes that while the open-source LIMS is quite usable as is, it will require further development of more "generic and configurable workflows." Despite this, the authors anticipate the software to be useful to the biobanking community.
This journal article, published in Biopreservation and Biobanking in early 2017, presents the development philosophy and implementation of a custom-modified version of Bika LIMS called Baobab LIMS, designed for biobank clients and researchers. Bendou et al., who enlisted customization help directly from Bika Lab System, describe how "[t]he need to implement biobank standard operation procedures as well as stimulate the use of standards for biobank data representation motivated the implementation of Baobab LIMS, an open-source LIMS for biobanking." The group concludes that while the open-source LIMS is quite usable as is, it will require further development of more "generic and configurable workflows." Despite this, the authors anticipate the software to be useful to the biobanking community.
 The problem? Disparate data sources, from weather and wave forecasts to navigation charts and natural hazard assessments, made oceanography research in Southern Italy more cumbersome. Solution? Create a secure, standardized, and interoperable data platform that can merge all that and other information together into one powerful and easy-to-use platform. Thus the TESSA (Development of Technology for Situational Sea Awareness) program was born. D'Anca et al. discuss the creation and use of TESSA as a geospatial tool that merges real-time and archived data to help researchers in Southern Italy. The authors conclude that TESSA is "a valid prototype easily adopted to provide an efficient dissemination of maritime data and a consolidation of the management of operational oceanographic activities," even in other parts of the world.
The problem? Disparate data sources, from weather and wave forecasts to navigation charts and natural hazard assessments, made oceanography research in Southern Italy more cumbersome. Solution? Create a secure, standardized, and interoperable data platform that can merge all that and other information together into one powerful and easy-to-use platform. Thus the TESSA (Development of Technology for Situational Sea Awareness) program was born. D'Anca et al. discuss the creation and use of TESSA as a geospatial tool that merges real-time and archived data to help researchers in Southern Italy. The authors conclude that TESSA is "a valid prototype easily adopted to provide an efficient dissemination of maritime data and a consolidation of the management of operational oceanographic activities," even in other parts of the world.
 In development since at least the summer of 2009, the open-source MASTR-MS laboratory information management system (LIMS) was designed to better handle the data and metadata of metabolomics, the study of an entity's metabolites. In this 2017 paper published in Metabolomics, the development team of MASTR-MS discuss the current state of their LIMS, how it's being used, and what the future holds for it. They conclude by stating the software's "comprehensive functions and features enable researchers and facilities to effectively manage a wide range of different project and experimental data types, and it facilitate the mining of new and existing [metabolomic] datasets."
In development since at least the summer of 2009, the open-source MASTR-MS laboratory information management system (LIMS) was designed to better handle the data and metadata of metabolomics, the study of an entity's metabolites. In this 2017 paper published in Metabolomics, the development team of MASTR-MS discuss the current state of their LIMS, how it's being used, and what the future holds for it. They conclude by stating the software's "comprehensive functions and features enable researchers and facilities to effectively manage a wide range of different project and experimental data types, and it facilitate the mining of new and existing [metabolomic] datasets."
 When designing something as simple as a menu of laboratory tests into a piece of clinical software, it's relatively easy to not think of the ramifications of the contents of such a menu. In this 2017 article published in BMC Medical Informatics and Decision Making, Martins et al. argue that there are consequences to what's included in a laboratory test drop-down menu, primarily that the presence — or lack thereof — of a test type may influence how frequently that test is prescribed. The group concludes that "[r]emoving unnecessary tests from a quick shortcut menu of the diagnosis and laboratory tests ordering system had a significant impact and reduced unnecessary prescription of tests," which in turn led "to the reduction of negative patient effects and to the reduction of unnecessary costs."
When designing something as simple as a menu of laboratory tests into a piece of clinical software, it's relatively easy to not think of the ramifications of the contents of such a menu. In this 2017 article published in BMC Medical Informatics and Decision Making, Martins et al. argue that there are consequences to what's included in a laboratory test drop-down menu, primarily that the presence — or lack thereof — of a test type may influence how frequently that test is prescribed. The group concludes that "[r]emoving unnecessary tests from a quick shortcut menu of the diagnosis and laboratory tests ordering system had a significant impact and reduced unnecessary prescription of tests," which in turn led "to the reduction of negative patient effects and to the reduction of unnecessary costs."
 In this journal article published in JMIR Medical Informatics in 2017, Alsaffar et al. review research from mid-2014 that looked at the state of open-source electronic health record (EHR) systems, primarily via SourceForge. The authors, noting a lack of research concerning the demographics and motivation of open-source EHR projects, present their finding, concluding that "lack of a corporate entity in most F/OSS EHR projects translates to a marginal capacity to market the respective F/OSS system and to navigate [HITECH] certification."
In this journal article published in JMIR Medical Informatics in 2017, Alsaffar et al. review research from mid-2014 that looked at the state of open-source electronic health record (EHR) systems, primarily via SourceForge. The authors, noting a lack of research concerning the demographics and motivation of open-source EHR projects, present their finding, concluding that "lack of a corporate entity in most F/OSS EHR projects translates to a marginal capacity to market the respective F/OSS system and to navigate [HITECH] certification."
 In this 2017 paper published in BMC Bioinformatics, Eyal-Altman et al. explain the use and benefits of their KNIME-based cancer outcome analysis software PCM-SABRE (Precision Cancer Medicine - Survival Analysis Benchmarking, Reporting and Evaluation). The group demonstrates its effectiveness by reconstructing the previous work of Chou et al. and showing how the results necessitate the tool for better reproducibility. The researchers conclude that when used effectively, PCM-Sabre's "resulting pipeline can be shared with others in an intuitive yet executable way, which will improve, if adopted by other investigators, the comparability and interpretability of future works attempting to predict patient survival from gene expression data."
In this 2017 paper published in BMC Bioinformatics, Eyal-Altman et al. explain the use and benefits of their KNIME-based cancer outcome analysis software PCM-SABRE (Precision Cancer Medicine - Survival Analysis Benchmarking, Reporting and Evaluation). The group demonstrates its effectiveness by reconstructing the previous work of Chou et al. and showing how the results necessitate the tool for better reproducibility. The researchers conclude that when used effectively, PCM-Sabre's "resulting pipeline can be shared with others in an intuitive yet executable way, which will improve, if adopted by other investigators, the comparability and interpretability of future works attempting to predict patient survival from gene expression data."
 In yet another installment of PLOS Computational Biology's Ten Simple Rules series, Boland et al. of Columbia University and the Broad Institute of MIT and Harvard share their thoughts and experiences with multi-site collaborations and data sharing. The group provides practical tips for making data sharing easier and more successful, strengthening collaborations and the scientific process.
In yet another installment of PLOS Computational Biology's Ten Simple Rules series, Boland et al. of Columbia University and the Broad Institute of MIT and Harvard share their thoughts and experiences with multi-site collaborations and data sharing. The group provides practical tips for making data sharing easier and more successful, strengthening collaborations and the scientific process.














