Posted on November 27, 2017
By Shawn Douglas
Journal articles
This brief review article from early 2017 looks at the basic elements of public health informatics and addresses how they're implemented. Aziz compares paper-based surveillance systems with electronic systems, noting the various improvements and challenges that have come with transitioning to electronic surveillance. He also reviews how public health informatics is applied in the U.S. and other parts of the world, including Saudi Arabia. Aziz concludes that "[p]atients, healthcare professionals, and public health officials can all help in reshaping public health through the adoption of new information systems, the use of electronic methods for disease surveillance, and the reformation of outmoded processes."
Posted on November 21, 2017
By Shawn Douglas
Journal articles

In this technical note from the University of Iowa Hospitals and Clinics, Imborek
et al. walk through the benefits and challenges of evaluating, implementing, and assessing preferred name and gender identity into its various laboratory systems and workflows. As preferred name has been an important point "in providing inclusion toward a class of patients that have historically been disenfranchised from the healthcare system" — primarily transgender patients — awareness is increasing at many levels about not only the inclusion of preferred name but also the complications that arise from it. From customizing software to billing and coding issues, from blood donor issues to laboratory test challenges, the group concludes that despite the "major undertaking [that] required the combined effort of many people from different teams," the efforts provide new opportunities for the field of pathology.
Posted on November 15, 2017
By Shawn Douglas
Journal articles
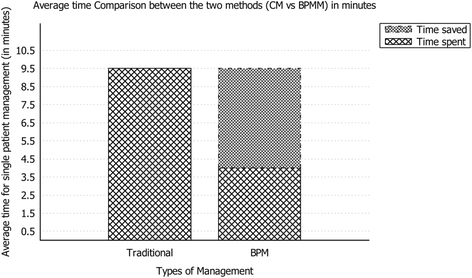
Sure, there's plenty of talk about big data in the research and clinical realms, but what of smarter data, gained from using business process management (BPM) tools? That's the direction Andellini
et al. went in after looking at the kidney transplantation cases at Bambino Gesù Children’s Hospital, realizing the potential for improving clinical decisions, scheduling of patient appointments, and reducing human errors in workflow. The group concluded "that the usage of BPM in the management of kidney transplantation leads to real benefits in terms of resource optimization and quality improvement," and that a "combination of a BPM-based platform with a service-oriented architecture could represent a revolution in clinical pathway management."
Posted on November 7, 2017
By Shawn Douglas
Journal articles
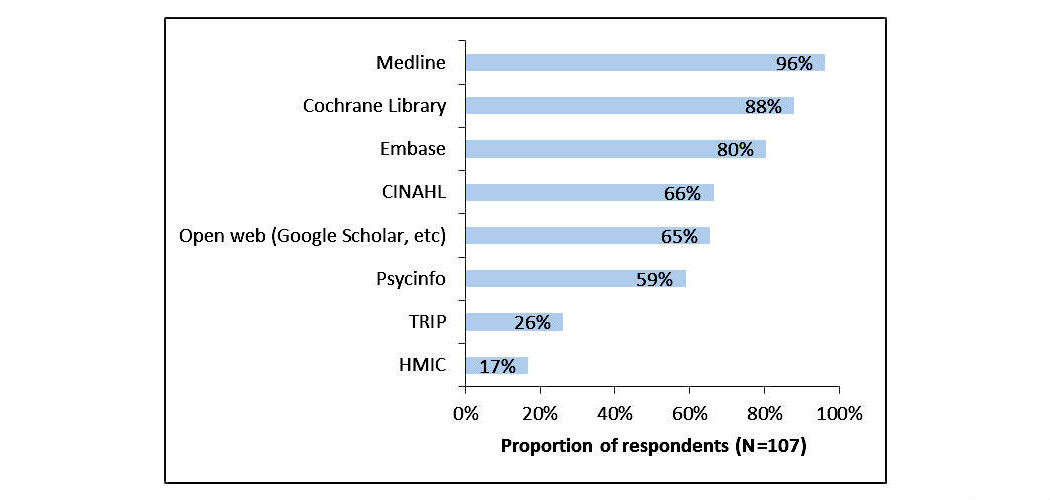
In this 2017 article published in
JMIR Medical Informatics, Russell-Rose and Chamberlain wished to take a closer look at the needs, goals, and requirements of healthcare information professionals performing complex searches of medical literature databases. Using a survey, they asked members of professional healthcare associations questions such as "how do you formulate search strategies?" and "what functionality do you value in a literature search system?". The researchers concluded that "current literature search systems offer only limited support for their requirements" and that "there is a need for improved functionality regarding the management of search strategies and the ability to search across multiple databases."
Posted on November 2, 2017
By Shawn Douglas
Journal articles
Unhappy with the electronic laboratory notebook (ELN) offerings for organic and inorganic chemists, a group of German researchers decided to take development of such an ELN into their own hands. This 2017 paper by Tremouilhac
et al. details the process of developing and implementing their own open-source solution, Chemotion ELN. After over a half year of use, the researchers state the ELN new serves "as a common infrastructure for chemistry research and [enables] chemistry researchers to build their own databases of digital information as a prerequisite for the detailed, systematic investigation and evaluation of chemical reactions and mechanisms." The group is hoping to add more functionality in time, including document generation and management, additional database query support, and an API for chemistry repositories.
Posted on October 24, 2017
By Shawn Douglas
Journal articles
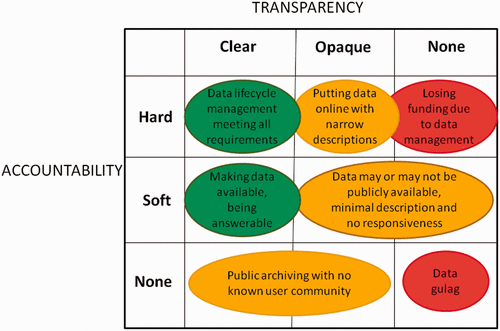
In this brief commentary article, University Corporation for Atmospheric Research's Matthew Mayernik discusses the concepts of "accountability" and "transparency" in regards to open data, conceptualizing open data "as the result of ongoing achievements, not one-time acts." In his conclusion, Mayernik notes that "good data management can happen even without sanctions," and that provision of access alone doesn't make something transparent, meaning that licenses and other complications make transparency difficult and require researchers "to account for changing expectations and requirements" of open data.
Posted on October 18, 2017
By Shawn Douglas
Journal articles
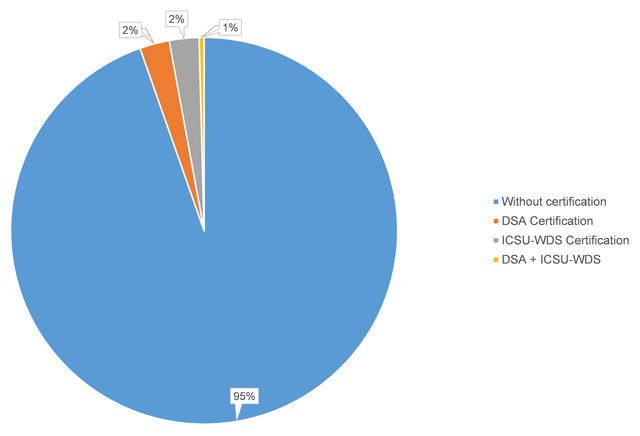
The use of publicly available data repositories is increasingly encouraged by organizations handling academic researchers' publications. Funding agencies, data organizations, and academic publishers alike are providing their researchers with lists of recommended repositories, though many aren't certified (as little as six percent of recommended are certified). Husen
et al. show that the landscape of recommended and certified data repositories is varied, and they conclude that "common ground for the standardization of data repository requirements" should be a common goal.
Posted on October 9, 2017
By Shawn Douglas
Journal articles
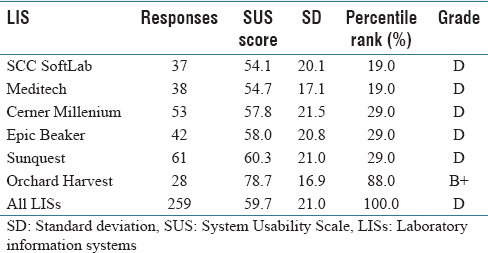
In this 2017 paper by Mathews and Marc, a picture is painted of laboratory information systems (LIS) usability, based on a structured survey with more than 250 qualifying responses: LIS design isn't particularly user-friendly. From
ad hoc laboratory reporting to quality control reporting, users consistently found several key LIS tasks either difficult or very difficult, with Orchard Harvest as the demonstrable exception. They conclude "that usability of an LIS is quite poor compared to various systems that have been evaluated using the [System Usability Scale]" though "[f]urther research is warranted to determine the root cause(s) of the difference in perceived usability" between Orchard Harvest and other discussed LIS.
Posted on October 3, 2017
By Shawn Douglas
Journal articles
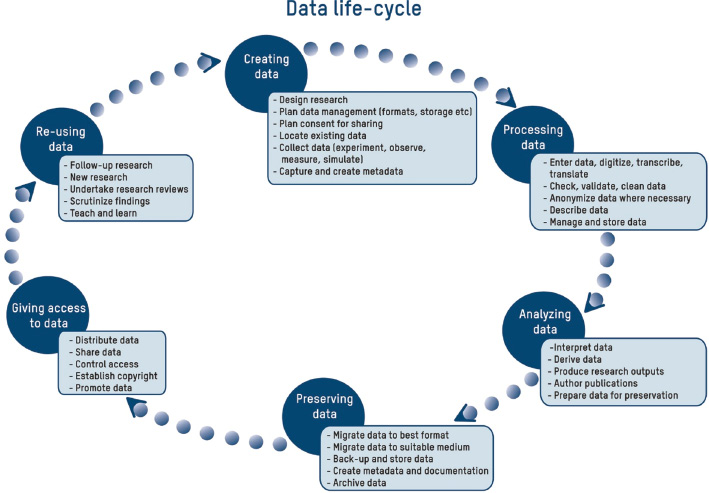
As calls for open scientific data get louder — particularly in Europe — research institutes, organizations, and data management stakeholders are being forced to consider their data management policies and research workflow in an attempt to better answer those calls. Martin
et al. of the National Research Institute of Science and Technology for Environment and Agriculture (Irstea) in France present their institute's efforts towards those goals in this 2017 paper published in
LIBER Quarterly. They refer frequently to the scientific and technical information (STI) professionals who must take on new skills, develop new policies, and implement new tools to better meet the goals of open data. Martin
et al. conclude with five points, foremost that these changes don't happen overnight; the necessary change "requires adaptation to technological developments and changes in scientific practices" in order to be successful.
Posted on September 26, 2017
By Shawn Douglas
Journal articles
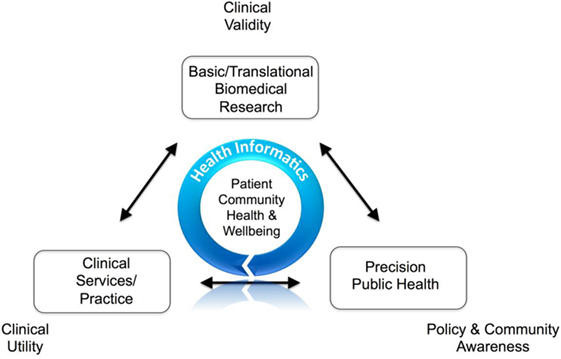
This short opinion article by Bellgard
et al. provides an Australian take on the role of information communication technology (ICT) in the healthcare and clinical research spheres. Despite the many opportunities for transforming those fields, the authors recognize that the successful use of ICT depends on a laundry list of factors, from data privacy policy to diverse stakeholder engagement. They conclude that ICT solutions for healthcare and clinical research "must be designed to be able to apply open data standards and open system principles that promote interoperability, service oriented architectures, application programming interfaces, and [allow for the] appropriate assessment of legacy ICT systems."
Posted on September 19, 2017
By Shawn Douglas
Journal articles
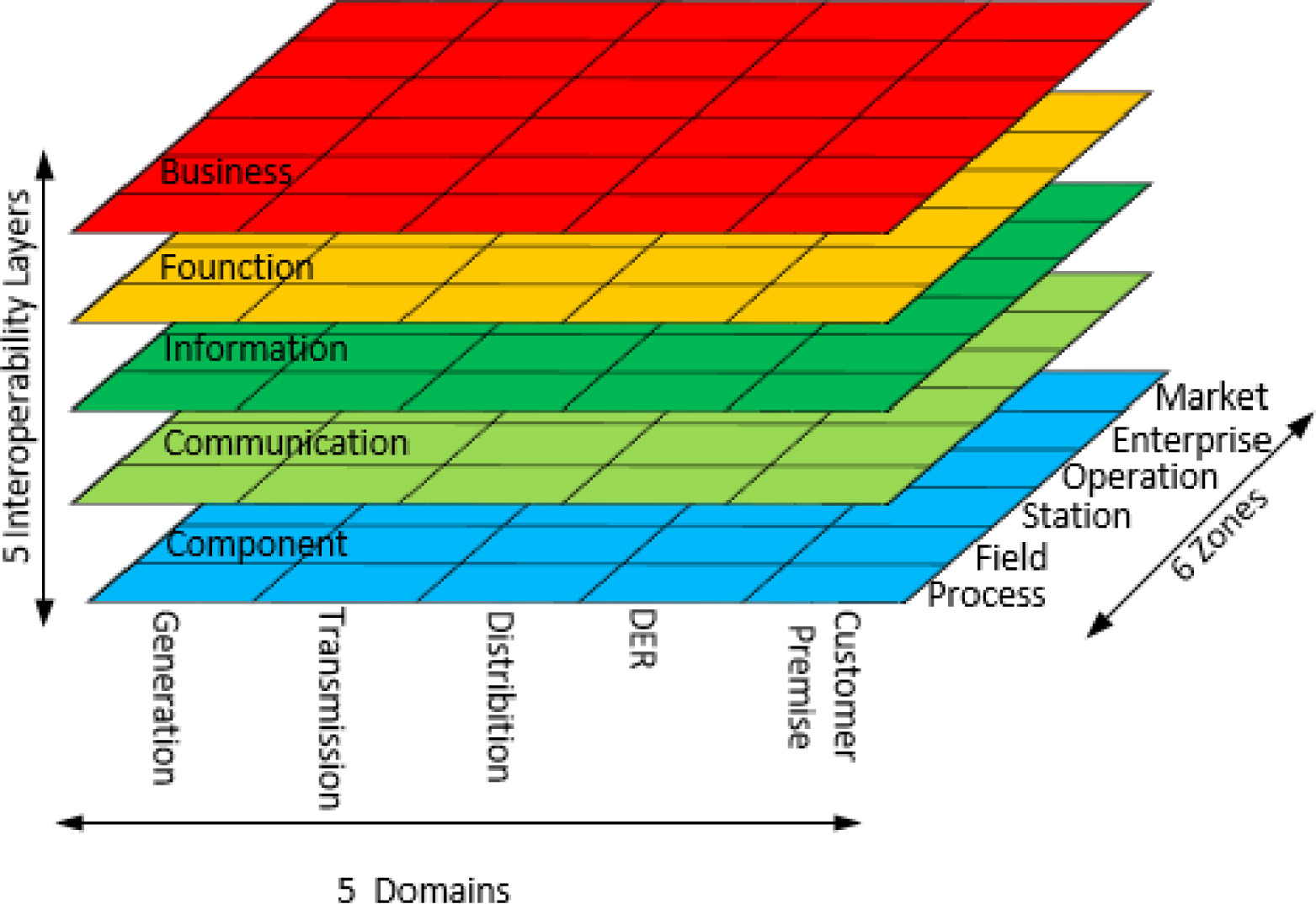
In this 2017 paper published in
ICT Express, Huang
et al., associated with a variety of power research institutes in China, provide an overview of energy informatics and the standardization efforts that have occurred around it and the concept of the "smart grid." After describing the various energy systems, technical fundamentals, and standardization efforts, the group concludes that "[l]earning from the successful concepts and technologies pioneered by the internet, and establishing an open interconnection protocol are basic methods for the successful development of the future energy system." They also add that "[i]t is essential to build an intelligent information system (or platform) to promote the interoperability and coordination of different energy systems and [information and communication technology] planes."
Posted on September 12, 2017
By Shawn Douglas
Journal articles
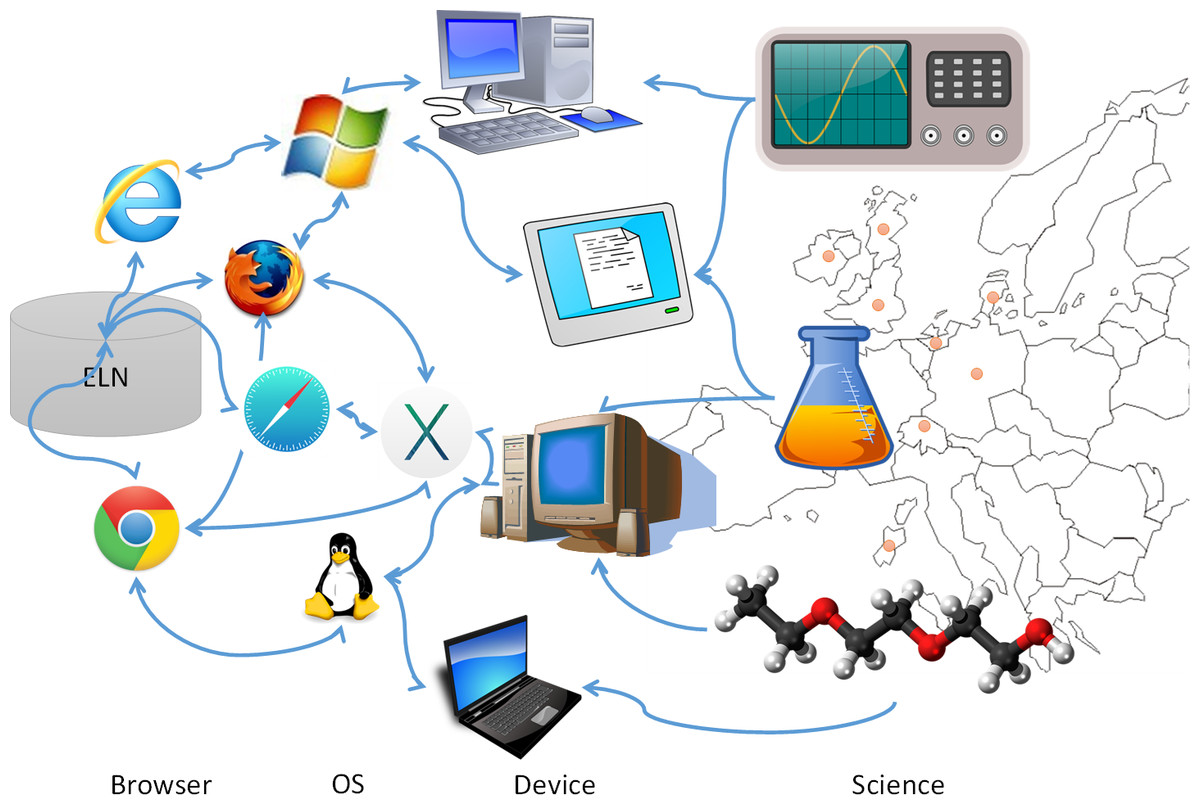
The electronic laboratory notebook (ELN) is implemented in a wide variety of research environments, but what are the special requirements of a public-private partnership? In this 2016 paper published in
PeerJ Computer Science, members of the TRANSLOCATION consortium — "a multinational and multisite public–private partnership (PPP) with 15 academic partners, five pharmaceutical companies and seven small- and medium-sized enterprises" — carefully present the process they took in selecting, installing, supporting, and re-evaluating an ELN for their scientific research. The group concludes that selection, implementation, change management. user buy-in, and value-added ability are all vital to the adoption and use of an ELN in a PPP.
Posted on September 6, 2017
By Shawn Douglas
Journal articles
This paper published in
Journal of Medical Biochemistry looks back on the benefits and errors associated with the implementation of a laboratory information system (LIS) in the Railway Healthcare Institute of Belgrade, Serbia. Author Vera Lukić explains how their first implementation went wrong and how a new LIS quickly helped improve the workflow of Railway's lab. Lukić finds that for them system flexibility and the ability to customize to user needs was most important in their implementation. He concludes that their LIS benefited them through the "increased pace of patient admission, prevention of sample identification errors, prevention of test translation errors, permanent results storage in electronic form, prevention of billing errors, improved time savings and better staff organization," resulting in "a step forward towards optimization of the total testing process."
Posted on August 28, 2017
By Shawn Douglas
Journal articles

This brief viewpoint article by Deliberato
et al. looks at the state of note taking in electronic health records (EHRs) and proposes that EHR developers look to artificial intelligence (AI) components to improve note taking and other tasks in their software. Not only would AI improve note taking, they argue, but "AI would provide helpful suggestions to the user about what information is available and how it might influence the next course of action. AI could also function to emphasize or deemphasize certain elements of the record, based on previous results, external databases, and knowledge networks."
Posted on August 23, 2017
By Shawn Douglas
Journal articles
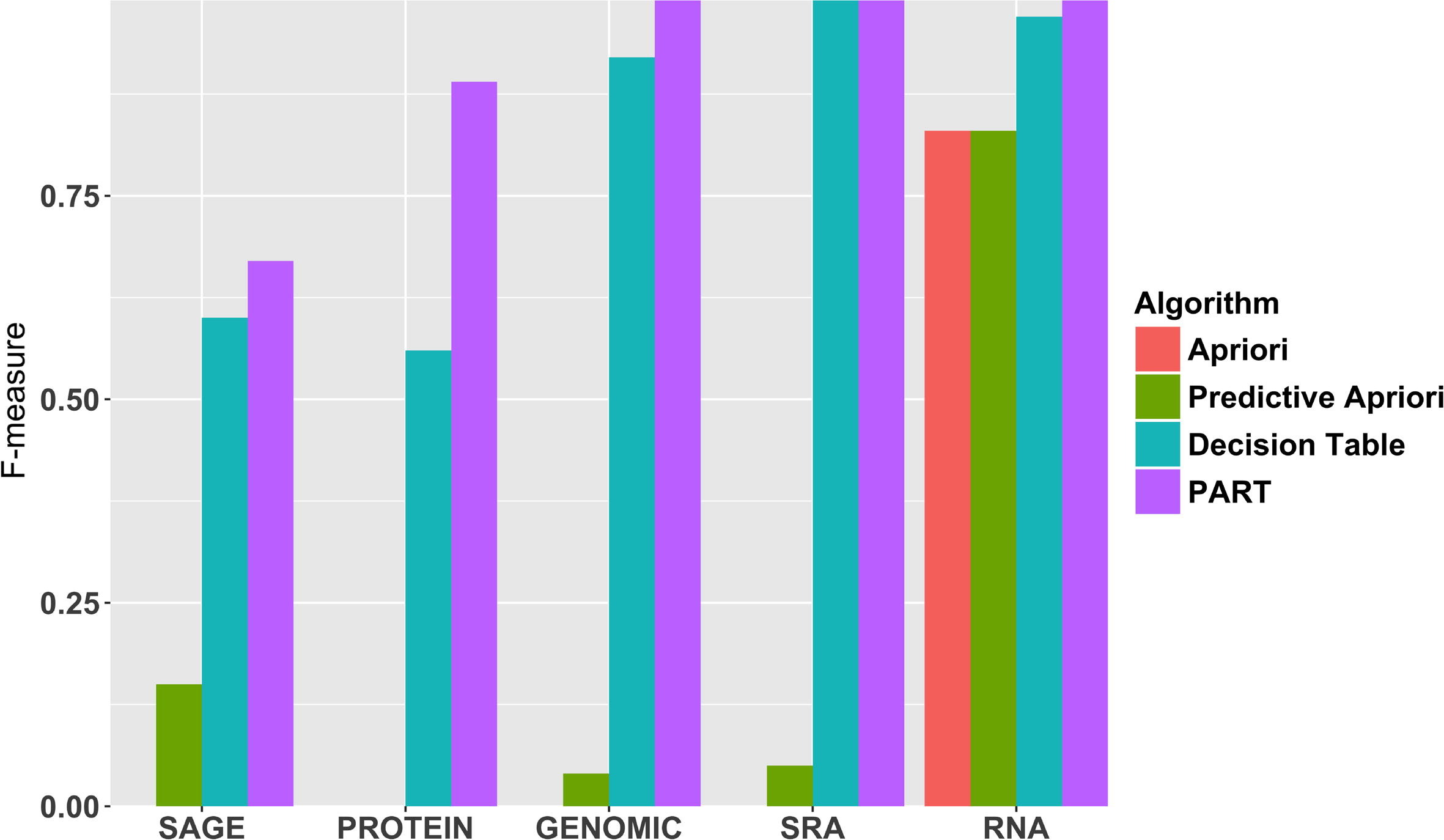
In this 2017 paper published in the
Journal of Biomedical Informatics, Panahiazar
et al. "propose a novel metadata prediction framework to learn associations from existing metadata that can be used to predict metadata values." They tested their framework using experimental metadata contained in the Gene Expression Omnibus (GEO), an international public repository for community-contributed genomic datasets. The framework itself uses association rule mining (ARM) algorithms to predict structured metadata, and they found that indeed ARM is a strong tool towards that goal, though with several limitations. They conclude that with tools like theirs, the discovered "[p]redictive metadata can be used both prospectively to facilitate metadata authoring, and retrospectively to improve, correct and augment existing metadata in biomedical databases."
Posted on August 16, 2017
By Shawn Douglas
Journal articles
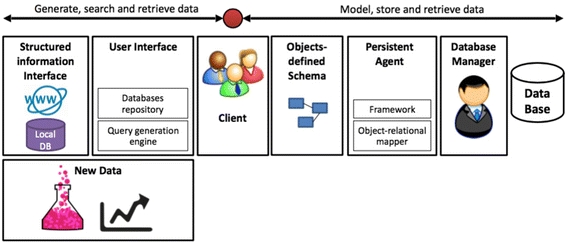
When it comes to bioinformatics data, databases are one of the more important the behind-the-scenes work horses. Yet inherent challenges of data heterogeneity and context-dependent interconnection in database design have driven the creation of specialized databases, which has, as a byproduct, caused additional problems in their creation. In this paper, Jerusalem College of Technology's Ezra Tsur proposes "an open-source framework for the curation of specialized databases," one that demonstrates "integration of the most relevant technologies to OO-based database design in a single framework" as well as extensibility to function with many other bioinformatics tools.
Posted on August 9, 2017
By Shawn Douglas
Journal articles
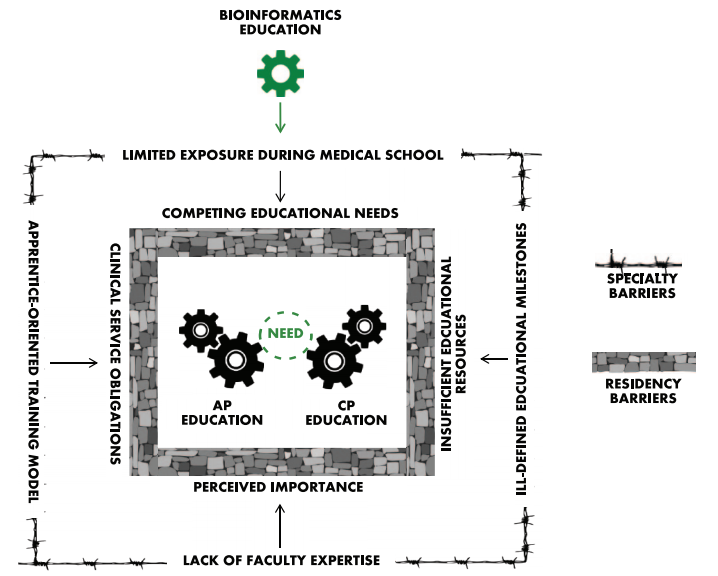
What are the links between pathology in the clinical setting and bioinformatics? Are residents in pathology gaining enough training in bioinformatics? And why should they learn about it in the first place? Clay and Fisher provide their take on these questions in this 2017 review paper published in
Cancer Informatics. From their point-of-view, bioinformatics training is vital to practicing pathology "in the 'information age' of diagnostic medicine," and training should be taken more seriously at the residency and fellowship levels. They conclude that "in order for bioinformatics education to firmly integrate into the fabric of resident education, its importance and broad application to the practice of pathology must be recognized and given a prominent seat at the education table."
Posted on August 2, 2017
By Shawn Douglas
Journal articles

This 2015 paper by Faria-Campos
et al. of the Brazilian Universidade Federal de Minas Gerais presents the reader with an overview of FluxCTTX, essentially a cytotoxicity module for the Flux laboratory information management system (LIMS). Citing a lack of laboratory informatics tools that can handle the specifics of cytotoxicity assays, the group develop FluxCTTX and tested it in five different laboratory environments, concluding that it can better "guarantee the quality of activities in the process of cytotoxicity tests and enforce the use of good laboratory practices (GLP)."
Posted on July 27, 2017
By Shawn Douglas
Journal articles
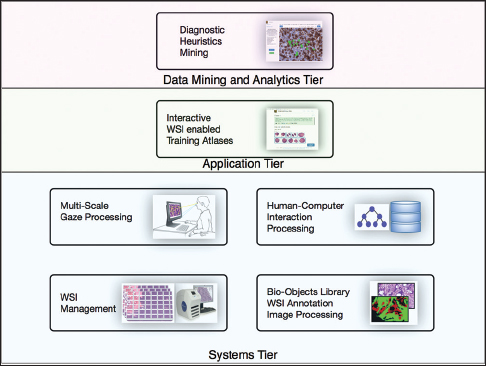
The visual and non-visual analysis techniques of pathology diagnosis are still in a relative infancy, representing a complex conglomeration of various data sources and techniques "which currently can be described more like a subjective exercise than a well-defined protocol." Enter Shin
et al., who developed the PathEdEx system, an informatics computational framework designed to pair digital pathology images and pathologists' eye/gaze patterns associated with those images to better understand and develop diagnostics methods and educational material for future pathologists. "All in all," they conclude, "the PathEdEx informatics tools have great potential to uncover, quantify, and study pathology diagnostic heuristics and can pave a path for precision diagnostics in the era of precision medicine."
Posted on July 26, 2017
By Shawn Douglas
Journal articles
Where are electronic laboratory notebooks (ELNs) going, and what do they lack? How does data from several user groups paint the picture of the ELN and its functionality and shortcomings? In this 2017 paper published in
Journal of Cheminformatics, researchers from the University of Southampton and BioSistemika examine the market and users of ELNs/paper laboratory notebooks, intent of identifying areas ELNs could be improved. They conclude that optimally there should be "an ELN environment that can serve as an interface between paper lab notebooks and the electronic documents that scientists create, one that is interoperable and utilizes semantic web and cloud technologies, particularly given that "current technology is such that it is desirable that ELN solutions work alongside paper for the foreseeable future."
 In this technical note from the University of Iowa Hospitals and Clinics, Imborek et al. walk through the benefits and challenges of evaluating, implementing, and assessing preferred name and gender identity into its various laboratory systems and workflows. As preferred name has been an important point "in providing inclusion toward a class of patients that have historically been disenfranchised from the healthcare system" — primarily transgender patients — awareness is increasing at many levels about not only the inclusion of preferred name but also the complications that arise from it. From customizing software to billing and coding issues, from blood donor issues to laboratory test challenges, the group concludes that despite the "major undertaking [that] required the combined effort of many people from different teams," the efforts provide new opportunities for the field of pathology.
In this technical note from the University of Iowa Hospitals and Clinics, Imborek et al. walk through the benefits and challenges of evaluating, implementing, and assessing preferred name and gender identity into its various laboratory systems and workflows. As preferred name has been an important point "in providing inclusion toward a class of patients that have historically been disenfranchised from the healthcare system" — primarily transgender patients — awareness is increasing at many levels about not only the inclusion of preferred name but also the complications that arise from it. From customizing software to billing and coding issues, from blood donor issues to laboratory test challenges, the group concludes that despite the "major undertaking [that] required the combined effort of many people from different teams," the efforts provide new opportunities for the field of pathology.
 Sure, there's plenty of talk about big data in the research and clinical realms, but what of smarter data, gained from using business process management (BPM) tools? That's the direction Andellini et al. went in after looking at the kidney transplantation cases at Bambino Gesù Children’s Hospital, realizing the potential for improving clinical decisions, scheduling of patient appointments, and reducing human errors in workflow. The group concluded "that the usage of BPM in the management of kidney transplantation leads to real benefits in terms of resource optimization and quality improvement," and that a "combination of a BPM-based platform with a service-oriented architecture could represent a revolution in clinical pathway management."
Sure, there's plenty of talk about big data in the research and clinical realms, but what of smarter data, gained from using business process management (BPM) tools? That's the direction Andellini et al. went in after looking at the kidney transplantation cases at Bambino Gesù Children’s Hospital, realizing the potential for improving clinical decisions, scheduling of patient appointments, and reducing human errors in workflow. The group concluded "that the usage of BPM in the management of kidney transplantation leads to real benefits in terms of resource optimization and quality improvement," and that a "combination of a BPM-based platform with a service-oriented architecture could represent a revolution in clinical pathway management."
 In this 2017 article published in JMIR Medical Informatics, Russell-Rose and Chamberlain wished to take a closer look at the needs, goals, and requirements of healthcare information professionals performing complex searches of medical literature databases. Using a survey, they asked members of professional healthcare associations questions such as "how do you formulate search strategies?" and "what functionality do you value in a literature search system?". The researchers concluded that "current literature search systems offer only limited support for their requirements" and that "there is a need for improved functionality regarding the management of search strategies and the ability to search across multiple databases."
In this 2017 article published in JMIR Medical Informatics, Russell-Rose and Chamberlain wished to take a closer look at the needs, goals, and requirements of healthcare information professionals performing complex searches of medical literature databases. Using a survey, they asked members of professional healthcare associations questions such as "how do you formulate search strategies?" and "what functionality do you value in a literature search system?". The researchers concluded that "current literature search systems offer only limited support for their requirements" and that "there is a need for improved functionality regarding the management of search strategies and the ability to search across multiple databases."
 Unhappy with the electronic laboratory notebook (ELN) offerings for organic and inorganic chemists, a group of German researchers decided to take development of such an ELN into their own hands. This 2017 paper by Tremouilhac et al. details the process of developing and implementing their own open-source solution, Chemotion ELN. After over a half year of use, the researchers state the ELN new serves "as a common infrastructure for chemistry research and [enables] chemistry researchers to build their own databases of digital information as a prerequisite for the detailed, systematic investigation and evaluation of chemical reactions and mechanisms." The group is hoping to add more functionality in time, including document generation and management, additional database query support, and an API for chemistry repositories.
Unhappy with the electronic laboratory notebook (ELN) offerings for organic and inorganic chemists, a group of German researchers decided to take development of such an ELN into their own hands. This 2017 paper by Tremouilhac et al. details the process of developing and implementing their own open-source solution, Chemotion ELN. After over a half year of use, the researchers state the ELN new serves "as a common infrastructure for chemistry research and [enables] chemistry researchers to build their own databases of digital information as a prerequisite for the detailed, systematic investigation and evaluation of chemical reactions and mechanisms." The group is hoping to add more functionality in time, including document generation and management, additional database query support, and an API for chemistry repositories.
 In this brief commentary article, University Corporation for Atmospheric Research's Matthew Mayernik discusses the concepts of "accountability" and "transparency" in regards to open data, conceptualizing open data "as the result of ongoing achievements, not one-time acts." In his conclusion, Mayernik notes that "good data management can happen even without sanctions," and that provision of access alone doesn't make something transparent, meaning that licenses and other complications make transparency difficult and require researchers "to account for changing expectations and requirements" of open data.
In this brief commentary article, University Corporation for Atmospheric Research's Matthew Mayernik discusses the concepts of "accountability" and "transparency" in regards to open data, conceptualizing open data "as the result of ongoing achievements, not one-time acts." In his conclusion, Mayernik notes that "good data management can happen even without sanctions," and that provision of access alone doesn't make something transparent, meaning that licenses and other complications make transparency difficult and require researchers "to account for changing expectations and requirements" of open data.
 The use of publicly available data repositories is increasingly encouraged by organizations handling academic researchers' publications. Funding agencies, data organizations, and academic publishers alike are providing their researchers with lists of recommended repositories, though many aren't certified (as little as six percent of recommended are certified). Husen et al. show that the landscape of recommended and certified data repositories is varied, and they conclude that "common ground for the standardization of data repository requirements" should be a common goal.
The use of publicly available data repositories is increasingly encouraged by organizations handling academic researchers' publications. Funding agencies, data organizations, and academic publishers alike are providing their researchers with lists of recommended repositories, though many aren't certified (as little as six percent of recommended are certified). Husen et al. show that the landscape of recommended and certified data repositories is varied, and they conclude that "common ground for the standardization of data repository requirements" should be a common goal.
 In this 2017 paper by Mathews and Marc, a picture is painted of laboratory information systems (LIS) usability, based on a structured survey with more than 250 qualifying responses: LIS design isn't particularly user-friendly. From ad hoc laboratory reporting to quality control reporting, users consistently found several key LIS tasks either difficult or very difficult, with Orchard Harvest as the demonstrable exception. They conclude "that usability of an LIS is quite poor compared to various systems that have been evaluated using the [System Usability Scale]" though "[f]urther research is warranted to determine the root cause(s) of the difference in perceived usability" between Orchard Harvest and other discussed LIS.
In this 2017 paper by Mathews and Marc, a picture is painted of laboratory information systems (LIS) usability, based on a structured survey with more than 250 qualifying responses: LIS design isn't particularly user-friendly. From ad hoc laboratory reporting to quality control reporting, users consistently found several key LIS tasks either difficult or very difficult, with Orchard Harvest as the demonstrable exception. They conclude "that usability of an LIS is quite poor compared to various systems that have been evaluated using the [System Usability Scale]" though "[f]urther research is warranted to determine the root cause(s) of the difference in perceived usability" between Orchard Harvest and other discussed LIS.
 As calls for open scientific data get louder — particularly in Europe — research institutes, organizations, and data management stakeholders are being forced to consider their data management policies and research workflow in an attempt to better answer those calls. Martin et al. of the National Research Institute of Science and Technology for Environment and Agriculture (Irstea) in France present their institute's efforts towards those goals in this 2017 paper published in LIBER Quarterly. They refer frequently to the scientific and technical information (STI) professionals who must take on new skills, develop new policies, and implement new tools to better meet the goals of open data. Martin et al. conclude with five points, foremost that these changes don't happen overnight; the necessary change "requires adaptation to technological developments and changes in scientific practices" in order to be successful.
As calls for open scientific data get louder — particularly in Europe — research institutes, organizations, and data management stakeholders are being forced to consider their data management policies and research workflow in an attempt to better answer those calls. Martin et al. of the National Research Institute of Science and Technology for Environment and Agriculture (Irstea) in France present their institute's efforts towards those goals in this 2017 paper published in LIBER Quarterly. They refer frequently to the scientific and technical information (STI) professionals who must take on new skills, develop new policies, and implement new tools to better meet the goals of open data. Martin et al. conclude with five points, foremost that these changes don't happen overnight; the necessary change "requires adaptation to technological developments and changes in scientific practices" in order to be successful.
 This short opinion article by Bellgard et al. provides an Australian take on the role of information communication technology (ICT) in the healthcare and clinical research spheres. Despite the many opportunities for transforming those fields, the authors recognize that the successful use of ICT depends on a laundry list of factors, from data privacy policy to diverse stakeholder engagement. They conclude that ICT solutions for healthcare and clinical research "must be designed to be able to apply open data standards and open system principles that promote interoperability, service oriented architectures, application programming interfaces, and [allow for the] appropriate assessment of legacy ICT systems."
This short opinion article by Bellgard et al. provides an Australian take on the role of information communication technology (ICT) in the healthcare and clinical research spheres. Despite the many opportunities for transforming those fields, the authors recognize that the successful use of ICT depends on a laundry list of factors, from data privacy policy to diverse stakeholder engagement. They conclude that ICT solutions for healthcare and clinical research "must be designed to be able to apply open data standards and open system principles that promote interoperability, service oriented architectures, application programming interfaces, and [allow for the] appropriate assessment of legacy ICT systems."
 In this 2017 paper published in ICT Express, Huang et al., associated with a variety of power research institutes in China, provide an overview of energy informatics and the standardization efforts that have occurred around it and the concept of the "smart grid." After describing the various energy systems, technical fundamentals, and standardization efforts, the group concludes that "[l]earning from the successful concepts and technologies pioneered by the internet, and establishing an open interconnection protocol are basic methods for the successful development of the future energy system." They also add that "[i]t is essential to build an intelligent information system (or platform) to promote the interoperability and coordination of different energy systems and [information and communication technology] planes."
In this 2017 paper published in ICT Express, Huang et al., associated with a variety of power research institutes in China, provide an overview of energy informatics and the standardization efforts that have occurred around it and the concept of the "smart grid." After describing the various energy systems, technical fundamentals, and standardization efforts, the group concludes that "[l]earning from the successful concepts and technologies pioneered by the internet, and establishing an open interconnection protocol are basic methods for the successful development of the future energy system." They also add that "[i]t is essential to build an intelligent information system (or platform) to promote the interoperability and coordination of different energy systems and [information and communication technology] planes."
 The electronic laboratory notebook (ELN) is implemented in a wide variety of research environments, but what are the special requirements of a public-private partnership? In this 2016 paper published in PeerJ Computer Science, members of the TRANSLOCATION consortium — "a multinational and multisite public–private partnership (PPP) with 15 academic partners, five pharmaceutical companies and seven small- and medium-sized enterprises" — carefully present the process they took in selecting, installing, supporting, and re-evaluating an ELN for their scientific research. The group concludes that selection, implementation, change management. user buy-in, and value-added ability are all vital to the adoption and use of an ELN in a PPP.
The electronic laboratory notebook (ELN) is implemented in a wide variety of research environments, but what are the special requirements of a public-private partnership? In this 2016 paper published in PeerJ Computer Science, members of the TRANSLOCATION consortium — "a multinational and multisite public–private partnership (PPP) with 15 academic partners, five pharmaceutical companies and seven small- and medium-sized enterprises" — carefully present the process they took in selecting, installing, supporting, and re-evaluating an ELN for their scientific research. The group concludes that selection, implementation, change management. user buy-in, and value-added ability are all vital to the adoption and use of an ELN in a PPP.
 This brief viewpoint article by Deliberato et al. looks at the state of note taking in electronic health records (EHRs) and proposes that EHR developers look to artificial intelligence (AI) components to improve note taking and other tasks in their software. Not only would AI improve note taking, they argue, but "AI would provide helpful suggestions to the user about what information is available and how it might influence the next course of action. AI could also function to emphasize or deemphasize certain elements of the record, based on previous results, external databases, and knowledge networks."
This brief viewpoint article by Deliberato et al. looks at the state of note taking in electronic health records (EHRs) and proposes that EHR developers look to artificial intelligence (AI) components to improve note taking and other tasks in their software. Not only would AI improve note taking, they argue, but "AI would provide helpful suggestions to the user about what information is available and how it might influence the next course of action. AI could also function to emphasize or deemphasize certain elements of the record, based on previous results, external databases, and knowledge networks."
 In this 2017 paper published in the Journal of Biomedical Informatics, Panahiazar et al. "propose a novel metadata prediction framework to learn associations from existing metadata that can be used to predict metadata values." They tested their framework using experimental metadata contained in the Gene Expression Omnibus (GEO), an international public repository for community-contributed genomic datasets. The framework itself uses association rule mining (ARM) algorithms to predict structured metadata, and they found that indeed ARM is a strong tool towards that goal, though with several limitations. They conclude that with tools like theirs, the discovered "[p]redictive metadata can be used both prospectively to facilitate metadata authoring, and retrospectively to improve, correct and augment existing metadata in biomedical databases."
In this 2017 paper published in the Journal of Biomedical Informatics, Panahiazar et al. "propose a novel metadata prediction framework to learn associations from existing metadata that can be used to predict metadata values." They tested their framework using experimental metadata contained in the Gene Expression Omnibus (GEO), an international public repository for community-contributed genomic datasets. The framework itself uses association rule mining (ARM) algorithms to predict structured metadata, and they found that indeed ARM is a strong tool towards that goal, though with several limitations. They conclude that with tools like theirs, the discovered "[p]redictive metadata can be used both prospectively to facilitate metadata authoring, and retrospectively to improve, correct and augment existing metadata in biomedical databases."
 When it comes to bioinformatics data, databases are one of the more important the behind-the-scenes work horses. Yet inherent challenges of data heterogeneity and context-dependent interconnection in database design have driven the creation of specialized databases, which has, as a byproduct, caused additional problems in their creation. In this paper, Jerusalem College of Technology's Ezra Tsur proposes "an open-source framework for the curation of specialized databases," one that demonstrates "integration of the most relevant technologies to OO-based database design in a single framework" as well as extensibility to function with many other bioinformatics tools.
When it comes to bioinformatics data, databases are one of the more important the behind-the-scenes work horses. Yet inherent challenges of data heterogeneity and context-dependent interconnection in database design have driven the creation of specialized databases, which has, as a byproduct, caused additional problems in their creation. In this paper, Jerusalem College of Technology's Ezra Tsur proposes "an open-source framework for the curation of specialized databases," one that demonstrates "integration of the most relevant technologies to OO-based database design in a single framework" as well as extensibility to function with many other bioinformatics tools.
 What are the links between pathology in the clinical setting and bioinformatics? Are residents in pathology gaining enough training in bioinformatics? And why should they learn about it in the first place? Clay and Fisher provide their take on these questions in this 2017 review paper published in Cancer Informatics. From their point-of-view, bioinformatics training is vital to practicing pathology "in the 'information age' of diagnostic medicine," and training should be taken more seriously at the residency and fellowship levels. They conclude that "in order for bioinformatics education to firmly integrate into the fabric of resident education, its importance and broad application to the practice of pathology must be recognized and given a prominent seat at the education table."
What are the links between pathology in the clinical setting and bioinformatics? Are residents in pathology gaining enough training in bioinformatics? And why should they learn about it in the first place? Clay and Fisher provide their take on these questions in this 2017 review paper published in Cancer Informatics. From their point-of-view, bioinformatics training is vital to practicing pathology "in the 'information age' of diagnostic medicine," and training should be taken more seriously at the residency and fellowship levels. They conclude that "in order for bioinformatics education to firmly integrate into the fabric of resident education, its importance and broad application to the practice of pathology must be recognized and given a prominent seat at the education table."
 This 2015 paper by Faria-Campos et al. of the Brazilian Universidade Federal de Minas Gerais presents the reader with an overview of FluxCTTX, essentially a cytotoxicity module for the Flux laboratory information management system (LIMS). Citing a lack of laboratory informatics tools that can handle the specifics of cytotoxicity assays, the group develop FluxCTTX and tested it in five different laboratory environments, concluding that it can better "guarantee the quality of activities in the process of cytotoxicity tests and enforce the use of good laboratory practices (GLP)."
This 2015 paper by Faria-Campos et al. of the Brazilian Universidade Federal de Minas Gerais presents the reader with an overview of FluxCTTX, essentially a cytotoxicity module for the Flux laboratory information management system (LIMS). Citing a lack of laboratory informatics tools that can handle the specifics of cytotoxicity assays, the group develop FluxCTTX and tested it in five different laboratory environments, concluding that it can better "guarantee the quality of activities in the process of cytotoxicity tests and enforce the use of good laboratory practices (GLP)."
 The visual and non-visual analysis techniques of pathology diagnosis are still in a relative infancy, representing a complex conglomeration of various data sources and techniques "which currently can be described more like a subjective exercise than a well-defined protocol." Enter Shin et al., who developed the PathEdEx system, an informatics computational framework designed to pair digital pathology images and pathologists' eye/gaze patterns associated with those images to better understand and develop diagnostics methods and educational material for future pathologists. "All in all," they conclude, "the PathEdEx informatics tools have great potential to uncover, quantify, and study pathology diagnostic heuristics and can pave a path for precision diagnostics in the era of precision medicine."
The visual and non-visual analysis techniques of pathology diagnosis are still in a relative infancy, representing a complex conglomeration of various data sources and techniques "which currently can be described more like a subjective exercise than a well-defined protocol." Enter Shin et al., who developed the PathEdEx system, an informatics computational framework designed to pair digital pathology images and pathologists' eye/gaze patterns associated with those images to better understand and develop diagnostics methods and educational material for future pathologists. "All in all," they conclude, "the PathEdEx informatics tools have great potential to uncover, quantify, and study pathology diagnostic heuristics and can pave a path for precision diagnostics in the era of precision medicine."
 Where are electronic laboratory notebooks (ELNs) going, and what do they lack? How does data from several user groups paint the picture of the ELN and its functionality and shortcomings? In this 2017 paper published in Journal of Cheminformatics, researchers from the University of Southampton and BioSistemika examine the market and users of ELNs/paper laboratory notebooks, intent of identifying areas ELNs could be improved. They conclude that optimally there should be "an ELN environment that can serve as an interface between paper lab notebooks and the electronic documents that scientists create, one that is interoperable and utilizes semantic web and cloud technologies, particularly given that "current technology is such that it is desirable that ELN solutions work alongside paper for the foreseeable future."
Where are electronic laboratory notebooks (ELNs) going, and what do they lack? How does data from several user groups paint the picture of the ELN and its functionality and shortcomings? In this 2017 paper published in Journal of Cheminformatics, researchers from the University of Southampton and BioSistemika examine the market and users of ELNs/paper laboratory notebooks, intent of identifying areas ELNs could be improved. They conclude that optimally there should be "an ELN environment that can serve as an interface between paper lab notebooks and the electronic documents that scientists create, one that is interoperable and utilizes semantic web and cloud technologies, particularly given that "current technology is such that it is desirable that ELN solutions work alongside paper for the foreseeable future."















